STRUCTURE OF FULL LENGTH HUMAN AMPK (NON-PHOSPHORYLATED AT T-LOOP) IN COMPLEX WITH A SMALL MOLECULE ACTIVATOR, A BENZIMIDAZOLE DERIVATIVE (991)
Xiao, B., Hubbard, J.A., Gamblin, S.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'-AMP-activated protein kinase catalytic subunit alpha-2 | A, D [auth C] | 552 | Homo sapiens | Mutation(s): 0 Gene Names: PRKAA2, AMPK, AMPK2 EC: 2.7.11.1 (PDB Primary Data), 2.7.11.27 (PDB Primary Data), 2.7.11.31 (PDB Primary Data) | 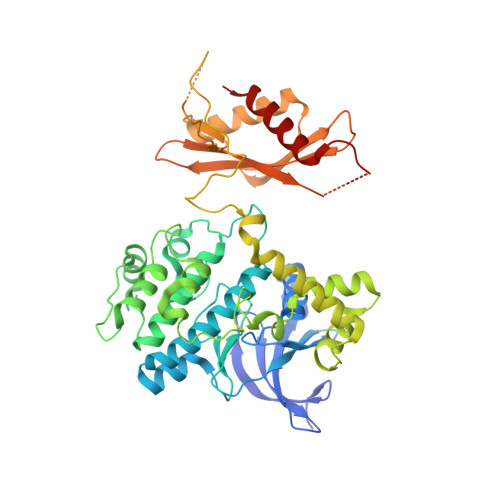 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P54646 (Homo sapiens) Explore P54646 Go to UniProtKB: P54646 | |||||
PHAROS: P54646 GTEx: ENSG00000162409 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P54646 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'-AMP-activated protein kinase subunit beta-1 | B, E [auth D] | 286 | Homo sapiens | Mutation(s): 0 Gene Names: PRKAB1, AMPK | 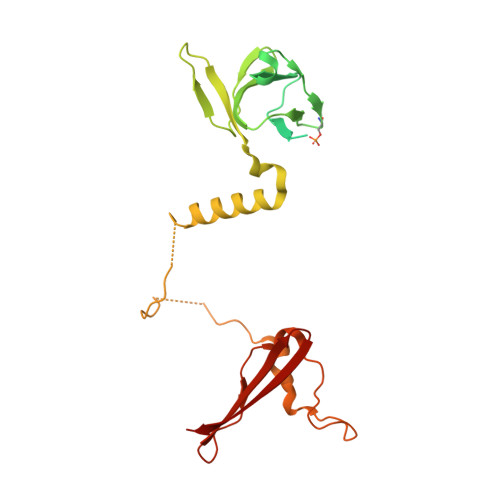 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y478 (Homo sapiens) Explore Q9Y478 Go to UniProtKB: Q9Y478 | |||||
PHAROS: Q9Y478 GTEx: ENSG00000111725 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y478 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 5'-AMP-activated protein kinase subunit gamma-1 | C [auth E], F | 331 | Homo sapiens | Mutation(s): 0 Gene Names: PRKAG1 | 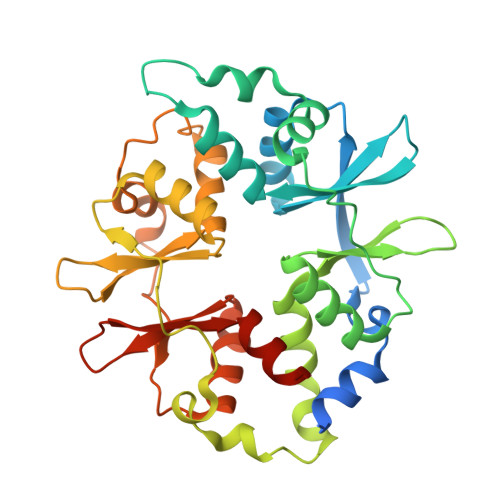 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P54619 (Homo sapiens) Explore P54619 Go to UniProtKB: P54619 | |||||
PHAROS: P54619 GTEx: ENSG00000181929 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P54619 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| STU Query on STU | G [auth A], L [auth C] | STAUROSPORINE C28 H26 N4 O3 HKSZLNNOFSGOKW-FYTWVXJKSA-N |  | ||
| 992 Query on 992 | H [auth A], M [auth C] | 5-[[6-chloranyl-5-(1-methylindol-5-yl)-1H-benzimidazol-2-yl]oxy]-2-methyl-benzoic acid C24 H18 Cl N3 O3 FHWSAZXFPUMKFL-UHFFFAOYSA-N |  | ||
| AMP Query on AMP | I [auth E] J [auth E] K [auth E] N [auth F] O [auth F] | ADENOSINE MONOPHOSPHATE C10 H14 N5 O7 P UDMBCSSLTHHNCD-KQYNXXCUSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | B, E [auth D] | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.42 | α = 90 |
| b = 129.39 | β = 92.73 |
| c = 139.28 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |