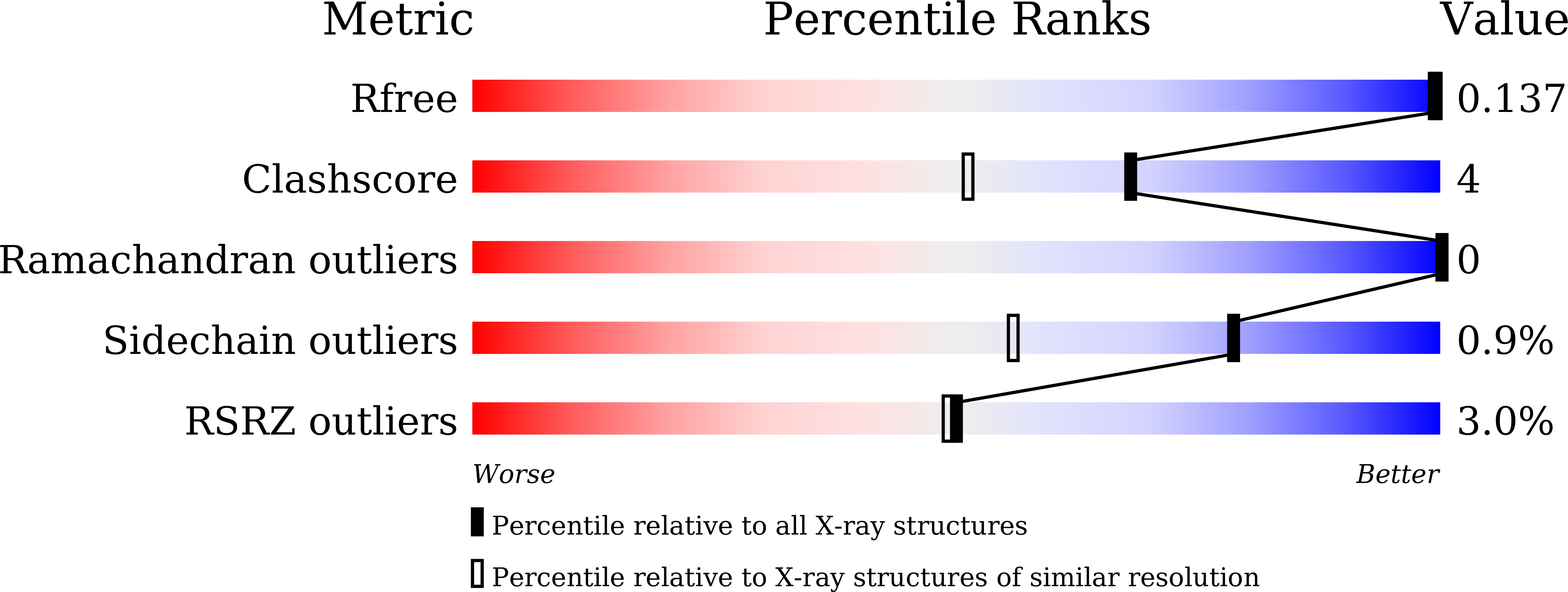
Structural basis for specific inhibition of the highly sensitive ShHTL7 receptor.
Shahul Hameed, U., Haider, I., Jamil, M., Kountche, B.A., Guo, X., Zarban, R.A., Kim, D., Al-Babili, S., Arold, S.T.(2018) EMBO Rep 19
- PubMed: 30021834
- DOI: https://doi.org/10.15252/embr.201745619
- Primary Citation of Related Structures:
5Z82, 5Z89, 5Z8P, 5Z95 - PubMed Abstract:
Striga hermonthica is a root parasitic plant that infests cereals, decimating yields, particularly in sub-Saharan Africa. For germination, Striga seeds require host-released strigolactones that are perceived by the family of HYPOSENSITIVE to LIGHT (ShHTL) receptors. Inhibiting seed germination would thus be a promising approach for combating Striga However, there are currently no strigolactone antagonists that specifically block ShHTLs and do not bind to DWARF14, the homologous strigolactone receptor of the host. Here, we show that the octyl phenol ethoxylate Triton X-100 inhibits S. hermonthica seed germination without affecting host plants. High-resolution X-ray structures reveal that Triton X-100 specifically plugs the catalytic pocket of ShHTL7. ShHTL7-specific inhibition by Triton X-100 demonstrates the dominant role of this particular ShHTL receptor for Striga germination. Our structural analysis provides a rationale for the broad specificity and high sensitivity of ShHTL7, and reveals that strigolactones trigger structural changes in ShHTL7 that are required for downstream signaling. Our findings identify Triton and the related 2-[4-(2,4,4-trimethylpentan-2-yl)phenoxy]acetic acid as promising lead compounds for the rational design of efficient Striga -specific herbicides.
Organizational Affiliation:
Division of Biological and Environmental Sciences and Engineering, Computational Bioscience Research Center, King Abdullah University of Science and Technology, Thuwal, Saudi Arabia.