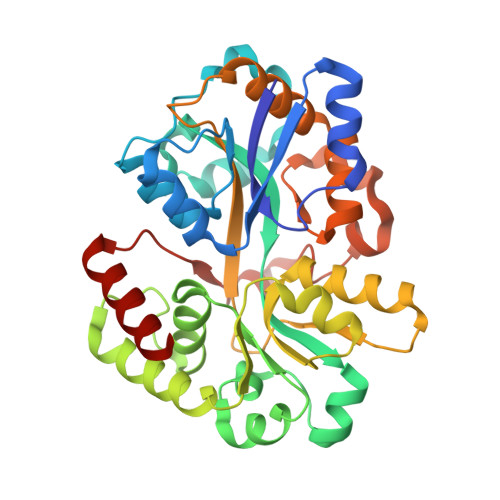
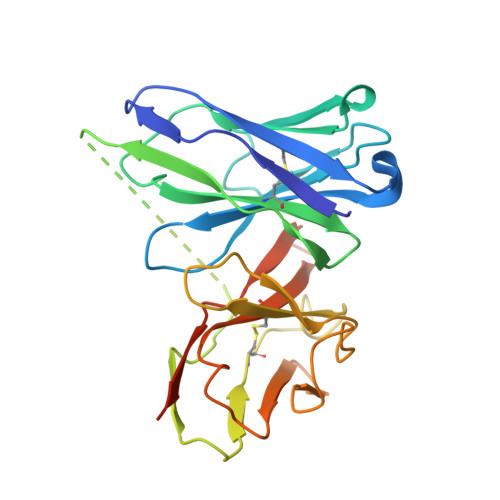
A Potent Anti-SpuE Antibody Allosterically Inhibits Type III Secretion System and Attenuates Virulence of Pseudomonas Aeruginosa.
Zhang, Y., Sun, X., Qian, Y., Yi, H., Song, K., Zhu, H., Zonta, F., Chen, W., Ji, Q., Miersch, S., Sidhu, S.S., Wu, D.(2019) J Mol Biol 431: 4882-4896
- PubMed: 31682834
- DOI: https://doi.org/10.1016/j.jmb.2019.10.026
- Primary Citation of Related Structures:
6IKM - PubMed Abstract:
Multidrug-resistant gram-negative bacteria infection is particularly severe within the designated ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), which underscores the urgent need to explore alternative therapeutic strategies. The type III secretion system (T3SS) is considered to be a key virulence factor in many gram-negative bacteria, and T3SS is in turn regulated by SpuE in P. aeruginosa, which is a spermidine binding protein from an ATP-binding cassette transporter family and highly conserved within ESKAPE pathogens. Here, we identified a potent anti-SpuE antagonistic antibody that allosterically inhibits the expression of T3SS and attenuates virulence of P. aeruginosa. X-ray crystallography and molecular dynamics simulations revealed that binding of antibody to SpuE induces a change in the dynamics of SpuE, which in turn may reduce spermidine uptake by P. aeruginosa. The antibody could serve as a template for developing novel biologics to target a broad spectrum of gram-negative bacteria.
Organizational Affiliation:
Laboratory of Antibody Engineering, Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Shanghai, China.