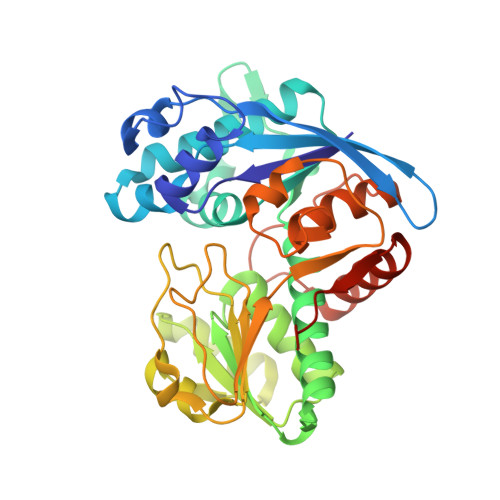
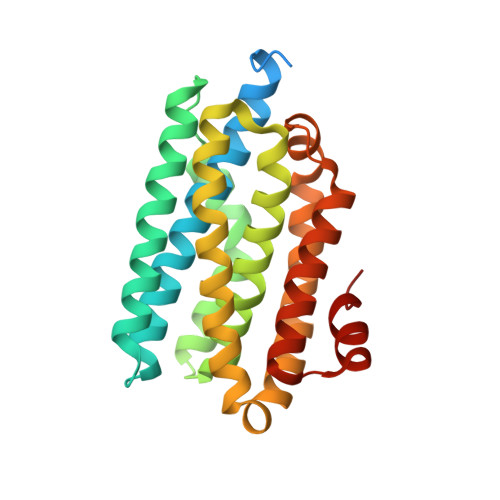
Structural insights into catalytic mechanism and product delivery of cyanobacterial acyl-acyl carrier protein reductase.
Gao, Y., Zhang, H., Fan, M., Jia, C., Shi, L., Pan, X., Cao, P., Zhao, X., Chang, W., Li, M.(2020) Nat Commun 11: 1525-1525
- PubMed: 32251275
- DOI: https://doi.org/10.1038/s41467-020-15268-y
- Primary Citation of Related Structures:
6JZQ, 6JZU, 6JZY, 6JZZ - PubMed Abstract:
Long-chain alk(a/e)nes represent the major constituents of conventional transportation fuels. Biosynthesis of alkanes is ubiquitous in many kinds of organisms. Cyanobacteria possess two enzymes, acyl-acyl carrier protein (acyl-ACP) reductase (AAR) and aldehyde-deformylating oxygenase (ADO), which function in a two-step alkane biosynthesis pathway. These two enzymes act in series and possibly form a complex that efficiently converts long chain fatty acyl-ACP/fatty acyl-CoA into hydrocarbon. While the structure of ADO has been previously described, structures of both AAR and AAR-ADO complex have not been solved, preventing deeper understanding of this pathway. Here, we report a ligand-free AAR structure, and three AAR-ADO complex structures in which AARs bind various ligands. Our results reveal the binding pattern of AAR with its substrate/cofactor, and suggest a potential aldehyde-transferring channel from AAR to ADO. Based on our structural and biochemical data, we proposed a model for the complete catalytic cycle of AAR.
- National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, People's Republic of China.
Organizational Affiliation: