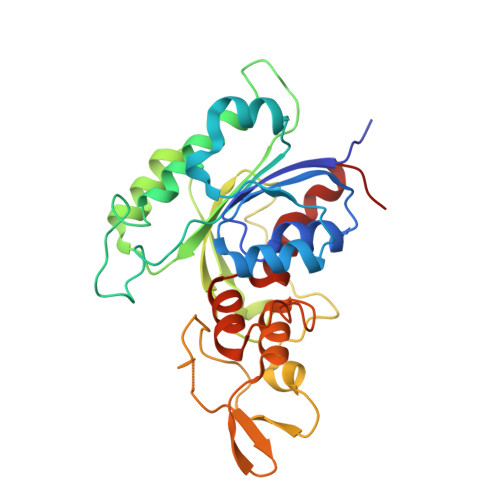
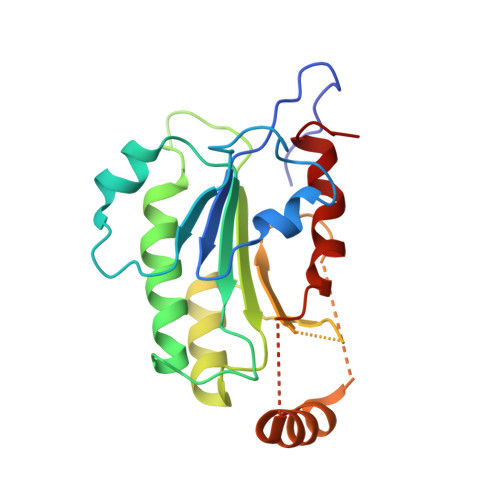
Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B.
Lin, C.C., Chen, Y.P., Yang, W.Z., Shen, J.C.K., Yuan, H.S.(2020) Nucleic Acids Res 48: 3949-3961
- PubMed: 32083663
- DOI: https://doi.org/10.1093/nar/gkaa111
- Primary Citation of Related Structures:
6KDA, 6KDB, 6KDL, 6KDP, 6KDT - PubMed Abstract:
DNA methyltransferases are primary enzymes for cytosine methylation at CpG sites of epigenetic gene regulation in mammals. De novo methyltransferases DNMT3A and DNMT3B create DNA methylation patterns during development, but how they differentially implement genomic DNA methylation patterns is poorly understood. Here, we report crystal structures of the catalytic domain of human DNMT3B-3L complex, noncovalently bound with and without DNA of different sequences. Human DNMT3B uses two flexible loops to enclose DNA and employs its catalytic loop to flip out the cytosine base. As opposed to DNMT3A, DNMT3B specifically recognizes DNA with CpGpG sites via residues Asn779 and Lys777 in its more stable and well-ordered target recognition domain loop to facilitate processive methylation of tandemly repeated CpG sites. We also identify a proton wire water channel for the final deprotonation step, revealing the complete working mechanism for cytosine methylation by DNMT3B and providing the structural basis for DNMT3B mutation-induced hypomethylation in immunodeficiency, centromere instability and facial anomalies syndrome.
- Institute of Molecular Biology, Academia Sinica, Taipei 11529, Taiwan.
Organizational Affiliation: