Odorant-binding site in visual opsin
Morizumi, T., Kuroi, K., Eger, B.T., Ou, W.L., Van Eps, N., Tsukamoto, H., Furutani, Y., Ernst, O.P.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Rhodopsin | 348 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes | 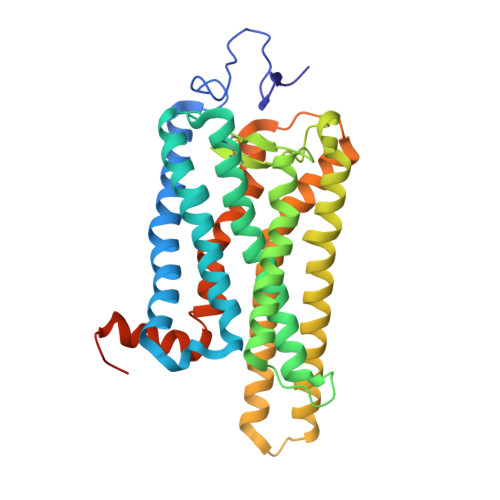 | |
UniProt | |||||
Find proteins for P02699 (Bos taurus) Explore P02699 Go to UniProtKB: P02699 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02699 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ILENLKDVGLF peptide CT2 | 11 | Bos taurus | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | C | 4 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G31886NL GlyCosmos: G31886NL GlyGen: G31886NL | |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BOG (Subject of Investigation/LOI) Query on BOG | D [auth A], E [auth A], F [auth A], G [auth A], K [auth A] | octyl beta-D-glucopyranoside C14 H28 O6 HEGSGKPQLMEBJL-RKQHYHRCSA-N |  | ||
| PLM Query on PLM | H [auth A] | PALMITIC ACID C16 H32 O2 IPCSVZSSVZVIGE-UHFFFAOYSA-N |  | ||
| SO4 Query on SO4 | I [auth A], J [auth A], L [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 242.169 | α = 90 |
| b = 242.169 | β = 90 |
| c = 110.716 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| autoPROC | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Canadian Institutes of Health Research (CIHR) | Canada | Canada Research Excellence Chair |