Identification and Optimization of EphA2-Selective Bicycles for the Delivery of Cytotoxic Payloads.
Mudd, G.E., Brown, A., Chen, L., van Rietschoten, K., Watcham, S., Teufel, D.P., Pavan, S., Lani, R., Huxley, P., Bennett, G.S.(2020) J Med Chem 63: 4107-4116
- PubMed: 32202781
- DOI: https://doi.org/10.1021/acs.jmedchem.9b02129
- Primary Citation of Related Structures:
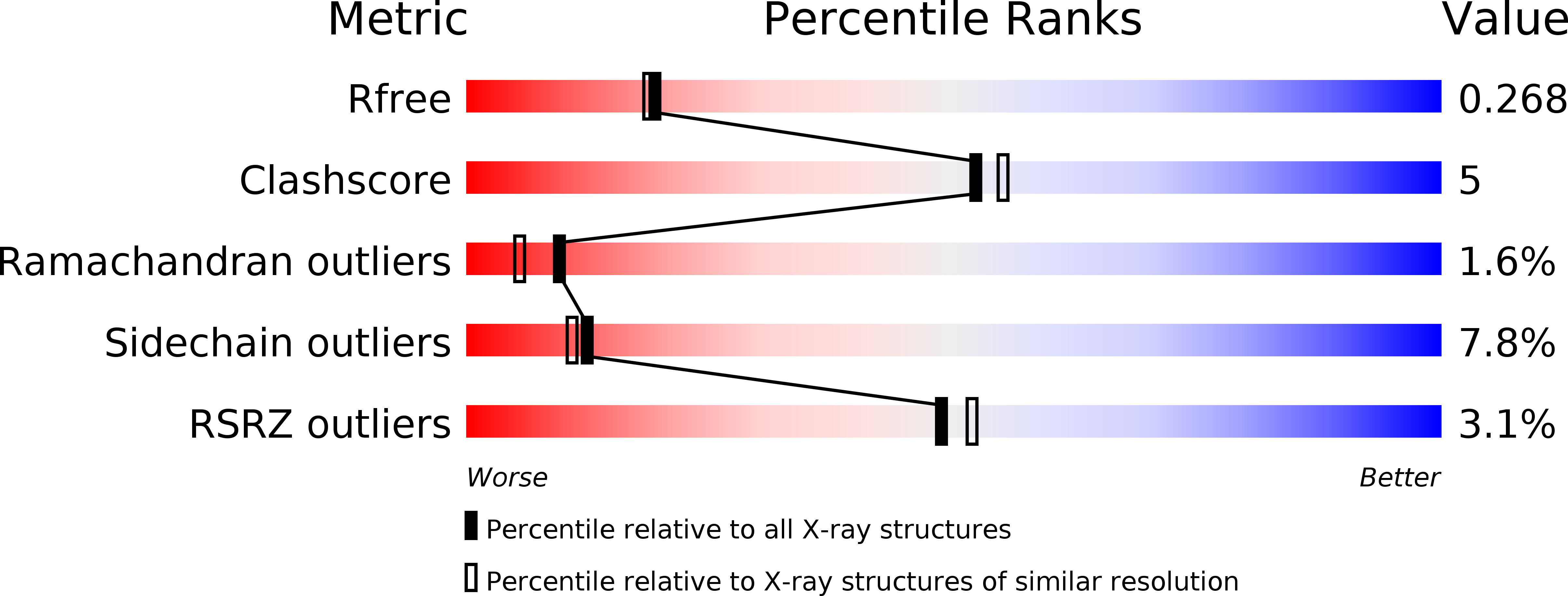
6RW2 - PubMed Abstract:
Bicycles are constrained bicyclic peptides that represent a promising binding modality for use in targeted drug conjugates. A phage display screen against EphA2, a receptor tyrosine kinase highly expressed in a number of solid tumors, identified a number of Bicycle families with low nanomolar affinity. A Bicycle toxin conjugate (BTC) was generated by derivatization of one of these Bicycles with the potent cytotoxin DM1 via a cleavable linker. This BTC demonstrated potent antitumor activity in vivo but was poorly tolerated, which was hypothesized to be the result of undesired liver uptake caused by poor physicochemical properties. Chemical optimization of a second Bicycle, guided by structural biology, provided a high affinity, metabolically stable Bicycle with improved physicochemical properties. A BTC incorporating this Bicycle also demonstrated potent antitumor activity and was very well tolerated when compared to the initial BTC. Phage display selection followed by chemical optimization of Bicycles can deliver potent drug conjugates with favorable pharmaceutical properties.
Organizational Affiliation:
BicycleTx Limited, Building 900, Babraham Research Campus, Cambridge CB22 3AT, United Kingdom.