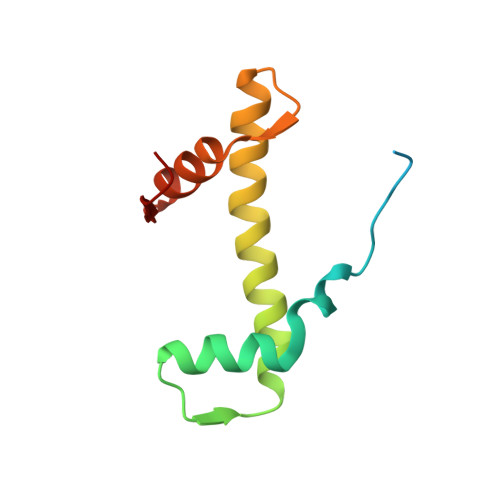
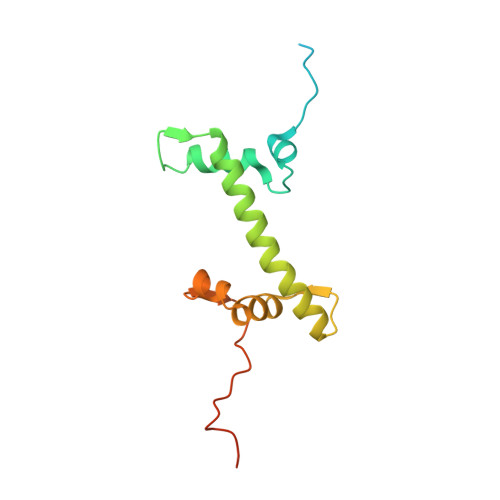
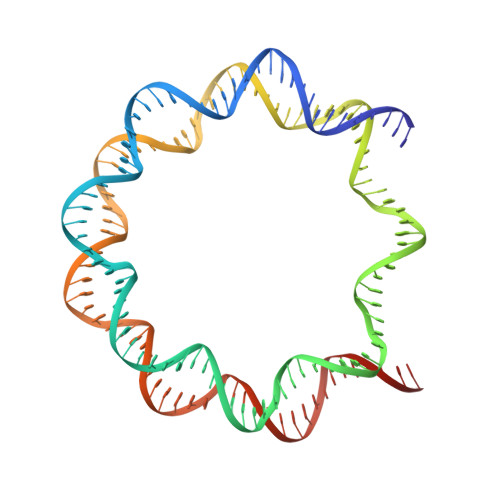
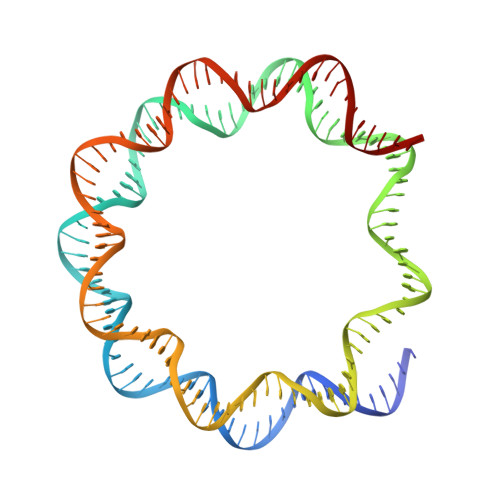
Nucleosome-bound SOX2 and SOX11 structures elucidate pioneer factor function.
Dodonova, S.O., Zhu, F., Dienemann, C., Taipale, J., Cramer, P.(2020) Nature 580: 669-672
- PubMed: 32350470
- DOI: https://doi.org/10.1038/s41586-020-2195-y
- Primary Citation of Related Structures:
6T78, 6T79, 6T7A, 6T7B, 6T7C, 6T7D - PubMed Abstract:
'Pioneer' transcription factors are required for stem-cell pluripotency, cell differentiation and cell reprogramming 1,2 . Pioneer factors can bind nucleosomal DNA to enable gene expression from regions of the genome with closed chromatin. SOX2 is a prominent pioneer factor that is essential for pluripotency and self-renewal of embryonic stem cells 3 . Here we report cryo-electron microscopy structures of the DNA-binding domains of SOX2 and its close homologue SOX11 bound to nucleosomes. The structures show that SOX factors can bind and locally distort DNA at superhelical location 2. The factors also facilitate detachment of terminal nucleosomal DNA from the histone octamer, which increases DNA accessibility. SOX-factor binding to the nucleosome can also lead to a repositioning of the N-terminal tail of histone H4 that includes residue lysine 16. We speculate that this repositioning is incompatible with higher-order nucleosome stacking, which involves contacts of the H4 tail with a neighbouring nucleosome. Our results indicate that pioneer transcription factors can use binding energy to initiate chromatin opening, and thereby facilitate nucleosome remodelling and subsequent transcription.
- Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany.
Organizational Affiliation: