The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification.
Houser, J., Kozmon, S., Mishra, D., Hammerova, Z., Wimmerova, M., Koca, J.(2020) Chemistry 26: 10769-10780
- PubMed: 32208534
- DOI: https://doi.org/10.1002/chem.202000593
- Primary Citation of Related Structures:
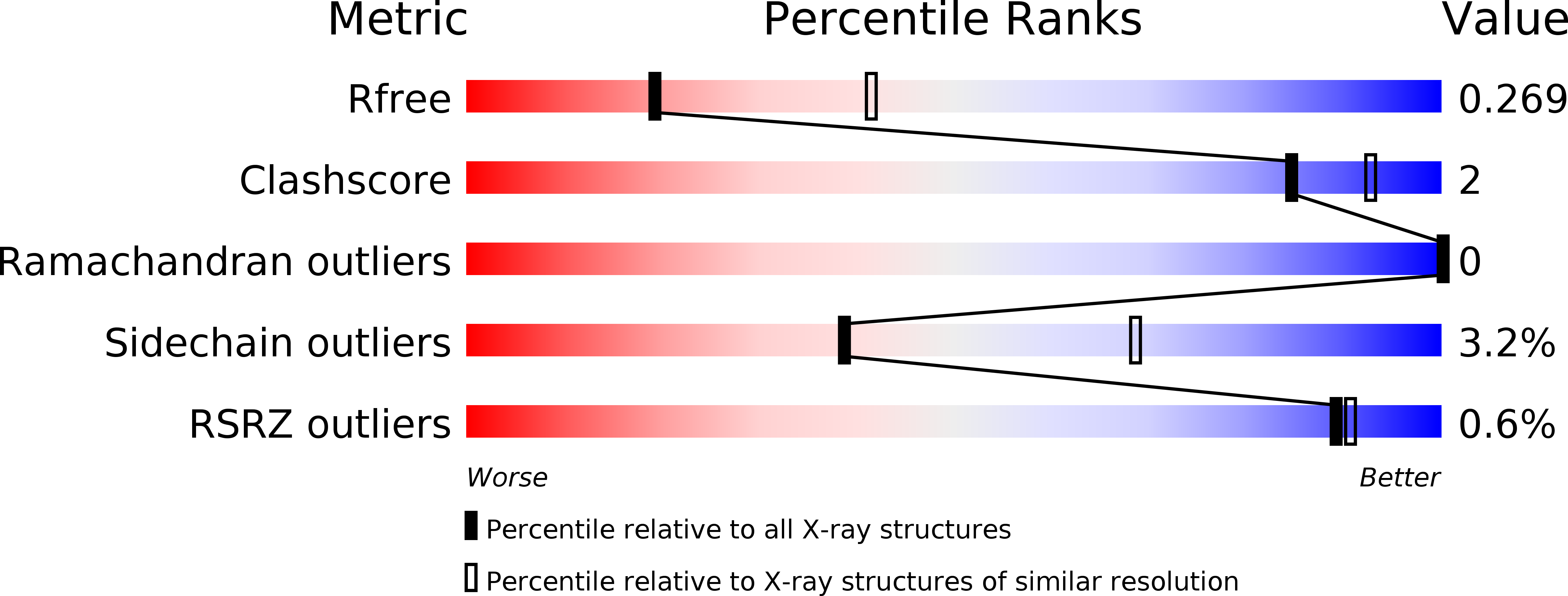
6T99, 6T9A, 6T9B - PubMed Abstract:
The molecular recognition of carbohydrates by proteins plays a key role in many biological processes including immune response, pathogen entry into a cell, and cell-cell adhesion (e.g., in cancer metastasis). Carbohydrates interact with proteins mainly through hydrogen bonding, metal-ion-mediated interaction, and non-polar dispersion interactions. The role of dispersion-driven CH-π interactions (stacking) in protein-carbohydrate recognition has been underestimated for a long time considering the polar interactions to be the main forces for saccharide interactions. However, over the last few years it turns out that non-polar interactions are equally important. In this study, we analyzed the CH-π interactions employing bioinformatics (data mining, structural analysis), several experimental (isothermal titration calorimetry (ITC), X-ray crystallography), and computational techniques. The Protein Data Bank (PDB) has been used as a source of structural data. The PDB contains over 12 000 protein complexes with carbohydrates. Stacking interactions are very frequently present in such complexes (about 39 % of identified structures). The calculations and the ITC measurement results suggest that the CH-π stacking contribution to the overall binding energy ranges from 4 up to 8 kcal mol -1 . All the results show that the stacking CH-π interactions in protein-carbohydrate complexes can be considered to be a driving force of the binding in such complexes.
Organizational Affiliation:
Central European Institute of Technology, Masaryk University, Kamenice 5, 62500, Brno, Czech Republic.