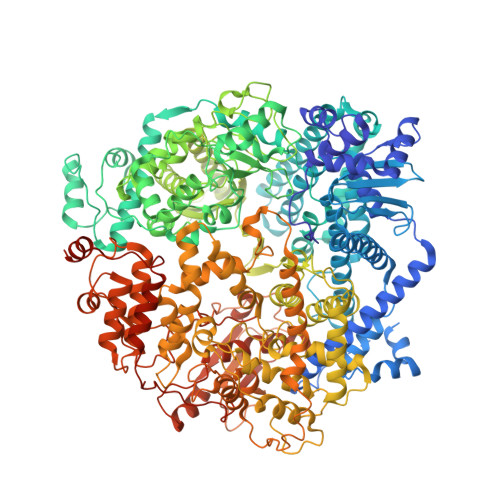
Cryo-EM structure of the respiratory syncytial virus RNA polymerase.
Cao, D., Gao, Y., Roesler, C., Rice, S., D'Cunha, P., Zhuang, L., Slack, J., Domke, M., Antonova, A., Romanelli, S., Keating, S., Forero, G., Juneja, P., Liang, B.(2020) Nat Commun 11: 368-368
- PubMed: 31953395
- DOI: https://doi.org/10.1038/s41467-019-14246-3
- Primary Citation of Related Structures:
6UEN - PubMed Abstract:
The respiratory syncytial virus (RSV) RNA polymerase, constituted of a 250 kDa large (L) protein and tetrameric phosphoprotein (P), catalyzes three distinct enzymatic activities - nucleotide polymerization, cap addition, and cap methylation. How RSV L and P coordinate these activities is poorly understood. Here, we present a 3.67 Å cryo-EM structure of the RSV polymerase (L:P) complex. The structure reveals that the RNA dependent RNA polymerase (RdRp) and capping (Cap) domains of L interact with the oligomerization domain (P OD ) and C-terminal domain (P CTD ) of a tetramer of P. The density of the methyltransferase (MT) domain of L and the N-terminal domain of P (P NTD ) is missing. Further analysis and comparison with other RNA polymerases at different stages suggest the structure we obtained is likely to be at an elongation-compatible stage. Together, these data provide enriched insights into the interrelationship, the inhibitors, and the evolutionary implications of the RSV polymerase.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA, 30322, USA.
Organizational Affiliation: