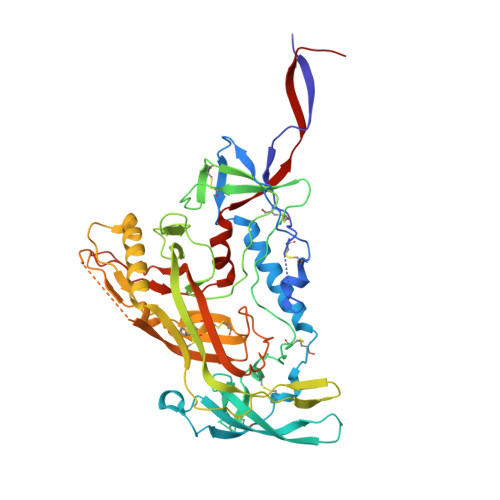
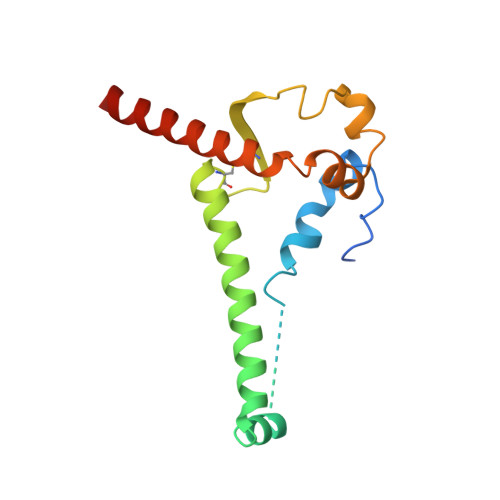
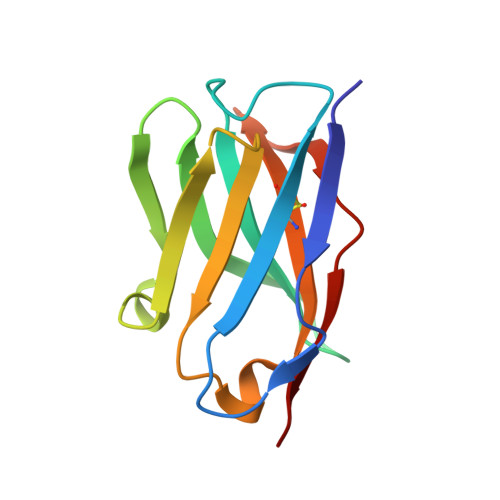
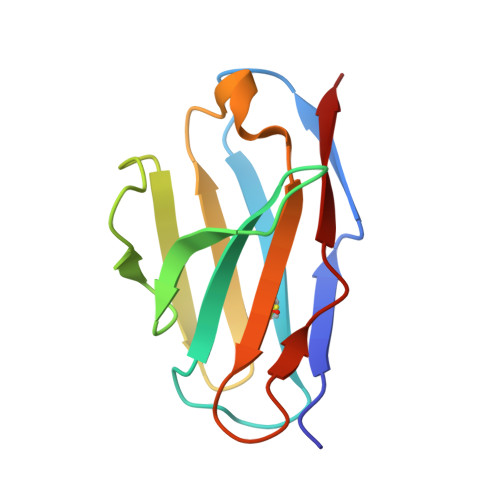Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Cottrell, C.A., van Schooten, J., Bowman, C.A., Yuan, M., Oyen, D., Shin, M., Morpurgo, R., van der Woude, P., van Breemen, M., Torres, J.L., Patel, R., Gross, J., Sewall, L.M., Copps, J., Ozorowski, G., Nogal, B., Sok, D., Rakasz, E.G., Labranche, C., Vigdorovich, V., Christley, S., Carnathan, D.G., Sather, D.N., Montefiori, D., Silvestri, G., Burton, D.R., Moore, J.P., Wilson, I.A., Sanders, R.W., Ward, A.B., van Gils, M.J.(2020) PLoS Pathog 16: e1008753-e1008753
- PubMed: 32866207
- DOI: https://doi.org/10.1371/journal.ppat.1008753
- Primary Citation of Related Structures:
6VLR, 6VN0, 6VO1, 6VOR, 6VOS, 6VSR - PubMed Abstract:
The induction of broad and potent immunity by vaccines is the key focus of research efforts aimed at protecting against HIV-1 infection. Soluble native-like HIV-1 envelope glycoproteins have shown promise as vaccine candidates as they can induce potent autologous neutralizing responses in rabbits and non-human primates. In this study, monoclonal antibodies were isolated and characterized from rhesus macaques immunized with the BG505 SOSIP.664 trimer to better understand vaccine-induced antibody responses. Our studies reveal a diverse landscape of antibodies recognizing immunodominant strain-specific epitopes and non-neutralizing neo-epitopes. Additionally, we isolated a subset of mAbs against an epitope cluster at the gp120-gp41 interface that recognize the highly conserved fusion peptide and the glycan at position 88 and have characteristics akin to several human-derived broadly neutralizing antibodies.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, United States of America.