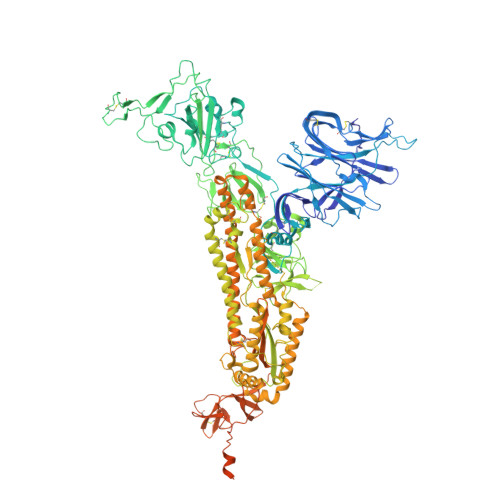
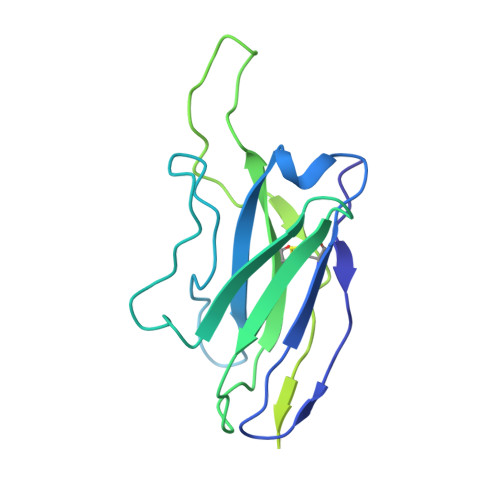
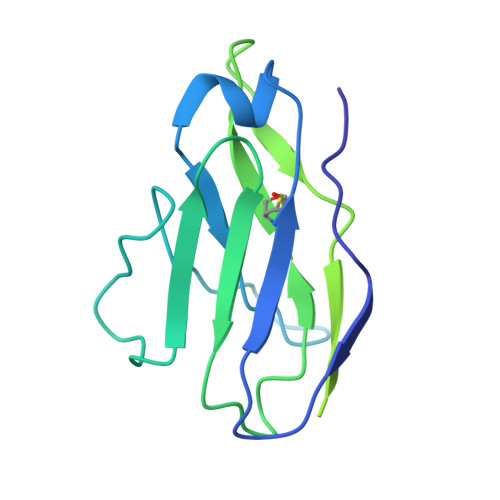
Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cerutti, G., Guo, Y., Zhou, T., Gorman, J., Lee, M., Rapp, M., Reddem, E.R., Yu, J., Bahna, F., Bimela, J., Huang, Y., Katsamba, P.S., Liu, L., Nair, M.S., Rawi, R., Olia, A.S., Wang, P., Zhang, B., Chuang, G.Y., Ho, D.D., Sheng, Z., Kwong, P.D., Shapiro, L.(2021) Cell Host Microbe 29: 819
- PubMed: 33789084
- DOI: https://doi.org/10.1016/j.chom.2021.03.005
- Primary Citation of Related Structures:
7L2C, 7L2D, 7L2E, 7L2F, 7LQW - PubMed Abstract:
Numerous antibodies that neutralize SARS-CoV-2 have been identified, and these generally target either the receptor-binding domain (RBD) or the N-terminal domain (NTD) of the viral spike. While RBD-directed antibodies have been extensively studied, far less is known about NTD-directed antibodies. Here, we report cryo-EM and crystal structures for seven potent NTD-directed neutralizing antibodies in complex with spike or isolated NTD. These structures defined several antibody classes, with at least one observed in multiple convalescent donors. The structures revealed that all seven antibodies target a common surface, bordered by glycans N17, N74, N122, and N149. This site-formed primarily by a mobile β-hairpin and several flexible loops-was highly electropositive, located at the periphery of the spike, and the largest glycan-free surface of NTD facing away from the viral membrane. Thus, in contrast to neutralizing RBD-directed antibodies that recognize multiple non-overlapping epitopes, potent NTD-directed neutralizing antibodies appear to target a single supersite.
- Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032, USA; Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY 10027, USA.
Organizational Affiliation: