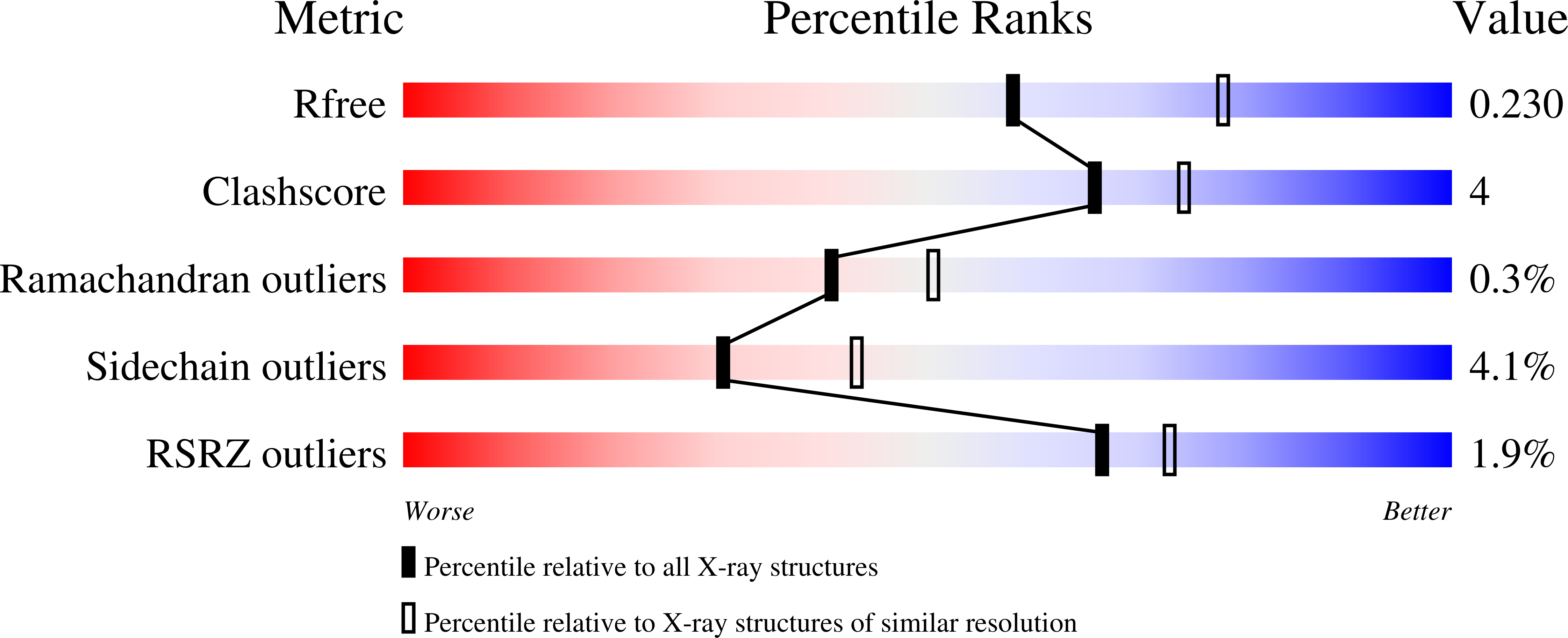
NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies.
Wagner, T.R., Ostertag, E., Kaiser, P.D., Gramlich, M., Ruetalo, N., Junker, D., Haering, J., Traenkle, B., Becker, M., Dulovic, A., Schweizer, H., Nueske, S., Scholz, A., Zeck, A., Schenke-Layland, K., Nelde, A., Strengert, M., Walz, J.S., Zocher, G., Stehle, T., Schindler, M., Schneiderhan-Marra, N., Rothbauer, U.(2021) EMBO Rep 22: e52325-e52325
- PubMed: 33904225
- DOI: https://doi.org/10.15252/embr.202052325
- Primary Citation of Related Structures:
7B27, 7NKT - PubMed Abstract:
In light of the COVID-19 pandemic, there is an ongoing need for diagnostic tools to monitor the immune status of large patient cohorts and the effectiveness of vaccination campaigns. Here, we present 11 unique nanobodies (Nbs) specific for the SARS-CoV-2 spike receptor-binding domain (RBD), of which 8 Nbs potently inhibit the interaction of RBD with angiotensin-converting enzyme 2 (ACE2) as the major viral docking site. Following detailed epitope mapping and structural analysis, we select two inhibitory Nbs, one of which binds an epitope inside and one of which binds an epitope outside the RBD:ACE2 interface. Based on these, we generate a biparatopic nanobody (bipNb) with viral neutralization efficacy in the picomolar range. Using bipNb as a surrogate, we establish a competitive multiplex binding assay ("NeutrobodyPlex") for detailed analysis of the presence and performance of neutralizing RBD-binding antibodies in serum of convalescent or vaccinated patients. We demonstrate that NeutrobodyPlex enables high-throughput screening and detailed analysis of neutralizing immune responses in infected or vaccinated individuals, to monitor immune status or to guide vaccine design.
Organizational Affiliation:
Pharmaceutical Biotechnology, Eberhard Karls University, Tuebingen, Germany.