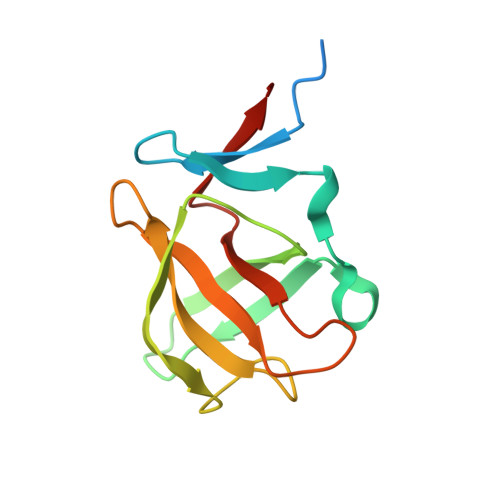
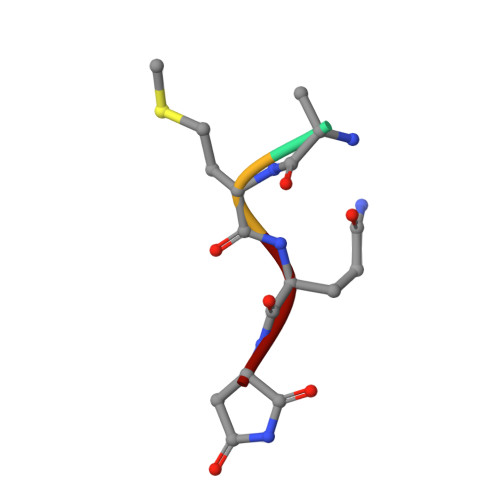
Identification and structural basis of C-terminal cyclic imides as natural degrons for cereblon.
Heim, C., Spring, A.K., Kirchgassner, S., Schwarzer, D., Hartmann, M.D.(2022) Biochem Biophys Res Commun 637: 66-72
- PubMed: 36375252
- DOI: https://doi.org/10.1016/j.bbrc.2022.11.001
- Primary Citation of Related Structures:
8BC6, 8BC7 - PubMed Abstract:
Cereblon (CRBN) is a ubiquitously expressed E3 ligase substrate receptor and a key player in pharmaceutical targeted protein degradation. Despite substantial insight gained into its chemical ligand space that is exploited in small-molecule protein degraders, its cellular role and native mechanism of substrate recognition remained elusive so far. In this communication, we report the discovery of C-terminal aspartimide and aminoglutarimide residues as natural degron motifs that are recognized by CRBN with high specificity. These C-terminal cyclic imides are known to form in ageing proteins as a result of spontaneous chain breaks after an attack of an asparagine or glutamine side chain amide on the adjacent peptide bond, and thereby mark potentially malfunctional protein fragments. In crystal structures, we uncover that these C-terminal cyclic imides are bound in the same fashion as small-molecule CRBN modulators, and that the residues preceding the cyclic terminus contribute to the interaction with a sequence-unspecific backbone hydrogen bonding pattern with strictly conserved residues in CRBN. We postulate that C-terminal aspartimide and aminoglutarimide residues resulting from chain breaks are largely underappreciated protein damages and represent the native degrons of CRBN.
- Max Planck Institute for Biology, Tübingen, Germany; Interfaculty Institute of Biochemistry, University of Tübingen, Tübingen, Germany; NanoTemper Technologies GmbH, Munich, Germany.
Organizational Affiliation: