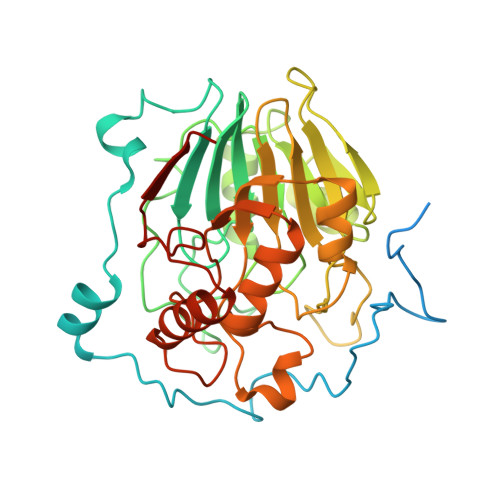
NAPE-PLD is target of thiazide diuretics.
Chiarugi, S., Margheriti, F., De Lorenzi, V., Martino, E., Margheritis, E.G., Moscardini, A., Marotta, R., Chaves-Sanjuan, A., Del Seppia, C., Federighi, G., Lapi, D., Bandiera, T., Rapposelli, S., Scuri, R., Bolognesi, M., Garau, G.(2025) Cell Chem Biol 32: 449
- PubMed: 39999832
- DOI: https://doi.org/10.1016/j.chembiol.2025.01.008
- Primary Citation of Related Structures:
8P90, 8P96, 8PC4 - PubMed Abstract:
Thiazide and thiazide-like diuretics are among the most efficacious and used drugs for the treatment of hypertension, edema, and major cardiovascular outcomes. Despite more then than six decades of clinical use, the molecular target and mechanism of action by which these drugs cure hypertension after long-term use have remained mysterious. Here we report the discovery and validation of a previously unknown renal and extrarenal target of these antihypertensives, the membrane-associated phospholipase N-acylphosphatidylethanolamine-specific phospholipase D (NAPE-PLD) of the endocannabinoid system. Structural and functional insights, together with preclinical studies in hypertensive rats, disclose the molecular and physiological basis by which thiazides cause acute diuresis and, at the same time, the distinctive chronic reduction of vascular resistance. Our results shed light on the mechanism of treatment of hypertension and will be useful for developing more efficacious medications for the management of vascular risk factors, as well as associated leukoencephalopathies and myelin disorders.
- BioStructures Lab, Istituto Italiano di Tecnologia (IIT@NEST), Piazza San Silvestro 12, 56127 Pisa, Italy; Laboratorio NEST, Scuola Normale Superiore, Piazza San Silvestro 12, 56127 Pisa, Italy.
Organizational Affiliation: