Automated Flow Peptide Synthesis Enables Engineering of Proteins with Stabilized Transient Binding Pockets.
Charalampidou, A., Nehls, T., Meyners, C., Gandhesiri, S., Pomplun, S., Pentelute, B.L., Lermyte, F., Hausch, F.(2024) ACS Cent Sci 10: 649-657
- PubMed: 38559286
- DOI: https://doi.org/10.1021/acscentsci.3c01283
- Primary Citation of Related Structures:
8PJ8, 8PJA - PubMed Abstract:
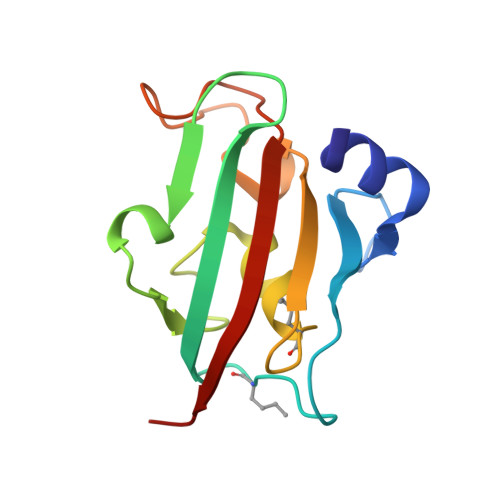
Engineering at the amino acid level is key to enhancing the properties of existing proteins in a desired manner. So far, protein engineering has been dominated by genetic approaches, which have been extremely powerful but only allow for minimal variations beyond the canonical amino acids. Chemical peptide synthesis allows the unrestricted incorporation of a vast set of unnatural amino acids with much broader functionalities, including the incorporation of post-translational modifications or labels. Here we demonstrate the potential of chemical synthesis to generate proteins in a specific conformation, which would have been unattainable by recombinant protein expression. We use recently established rapid automated flow peptide synthesis combined with solid-phase late-stage modifications to rapidly generate a set of FK506-binding protein 51 constructs bearing defined intramolecular lactam bridges. This trapped an otherwise rarely populated transient pocket-as confirmed by crystal structures-which led to an up to 39-fold improved binding affinity for conformation-selective ligands and represents a unique system for the development of ligands for this rare conformation. Overall, our results show how rapid automated flow peptide synthesis can be applied to precision protein engineering.
- Clemens-Schöpf-Institute, Department of Chemistry, Technical University of Darmstadt, Peter-Grünberg-Straße 4, 64287 Darmstadt, Germany.
Organizational Affiliation: