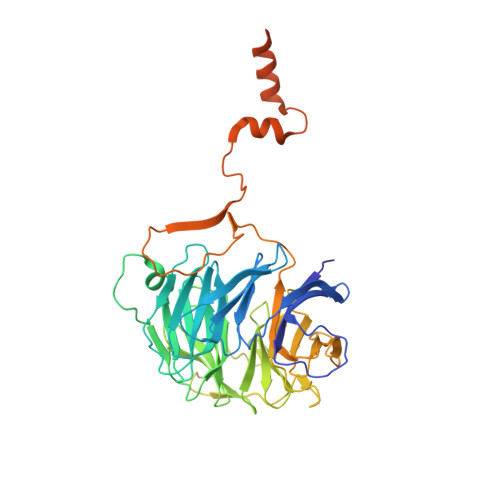
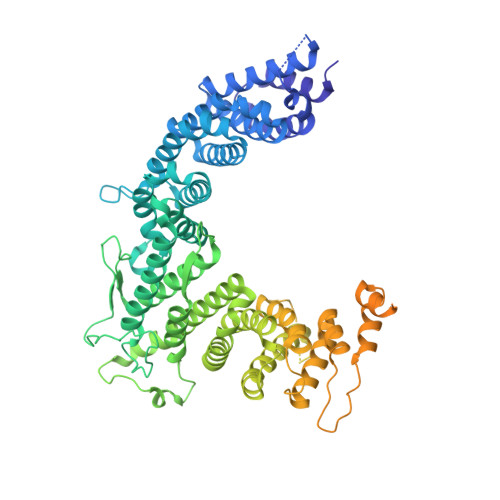
Structural insights into coordinating 5S RNP rotation with ITS2 pre-RNA processing during ribosome formation.
Thoms, M., Lau, B., Cheng, J., Fromm, L., Denk, T., Kellner, N., Flemming, D., Fischer, P., Falquet, L., Berninghausen, O., Beckmann, R., Hurt, E.(2023) EMBO Rep 24: e57984-e57984
- PubMed: 37921038
- DOI: https://doi.org/10.15252/embr.202357984
- Primary Citation of Related Structures:
8PTW, 8PUW, 8PV1, 8PV2, 8PV3, 8PV4, 8PV5, 8PV6, 8PV7, 8PV8, 8PVK, 8PVL - PubMed Abstract:
The rixosome defined in Schizosaccharomyces pombe and humans performs diverse roles in pre-ribosomal RNA processing and gene silencing. Here, we isolate and describe the conserved rixosome from Chaetomium thermophilum, which consists of two sub-modules, the sphere-like Rix1-Ipi3-Ipi1 and the butterfly-like Las1-Grc3 complex, connected by a flexible linker. The Rix1 complex of the rixosome utilizes Sda1 as landing platform on nucleoplasmic pre-60S particles to wedge between the 5S rRNA tip and L1-stalk, thereby facilitating the 180° rotation of the immature 5S RNP towards its mature conformation. Upon rixosome positioning, the other sub-module with Las1 endonuclease and Grc3 polynucleotide-kinase can reach a strategic position at the pre-60S foot to cleave and 5' phosphorylate the nearby ITS2 pre-rRNA. Finally, inward movement of the L1 stalk permits the flexible Nop53 N-terminus with its AIM motif to become positioned at the base of the L1-stalk to facilitate Mtr4 helicase-exosome participation for completing ITS2 removal. Thus, the rixosome structure elucidates the coordination of two central ribosome biogenesis events, but its role in gene silencing may adapt similar strategies.
- Gene Center, Ludwig-Maximilians-Universität München, Munich, Germany.
Organizational Affiliation: