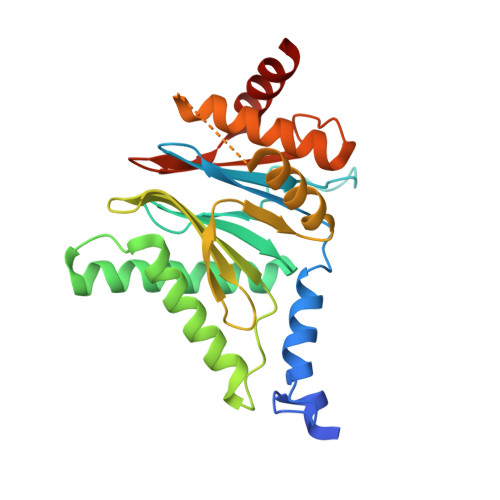
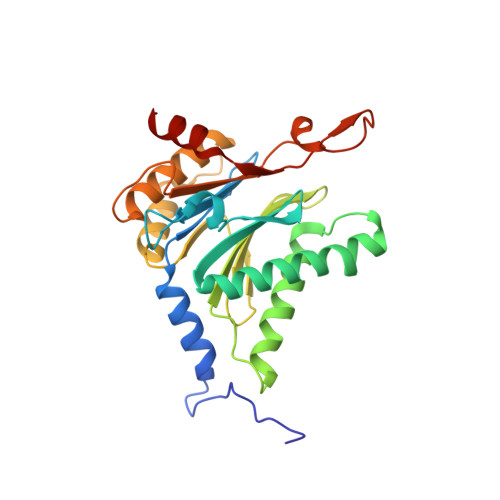
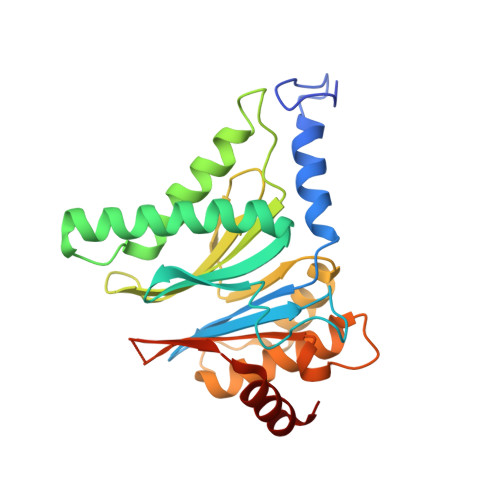
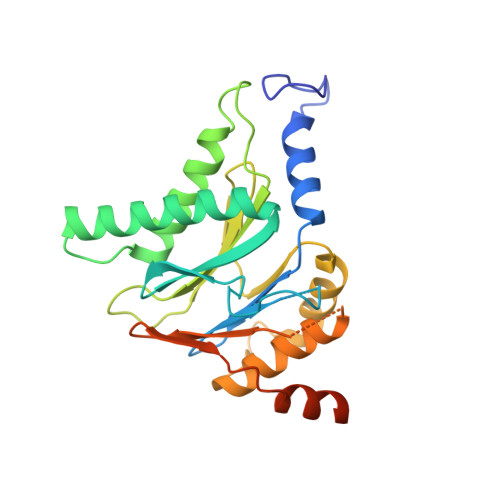
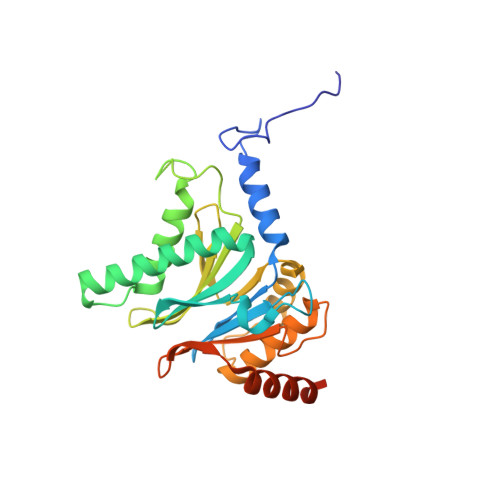
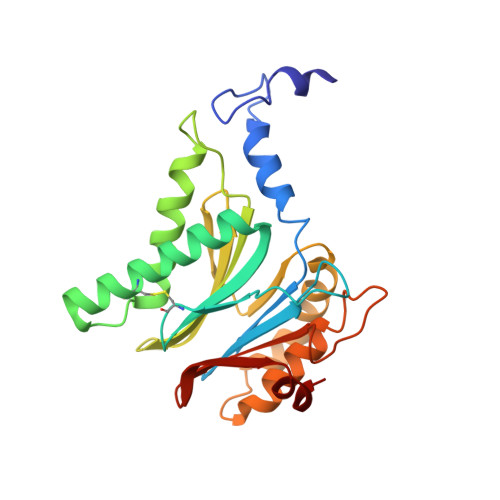
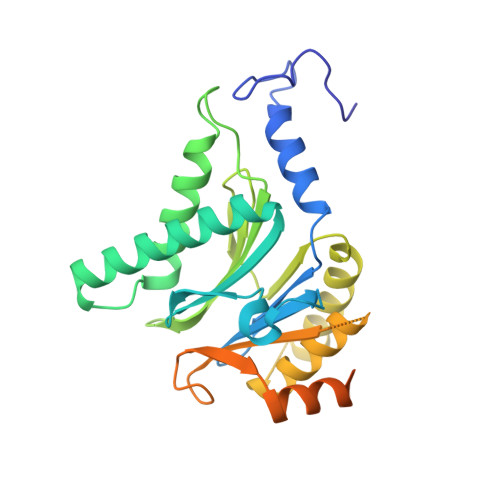
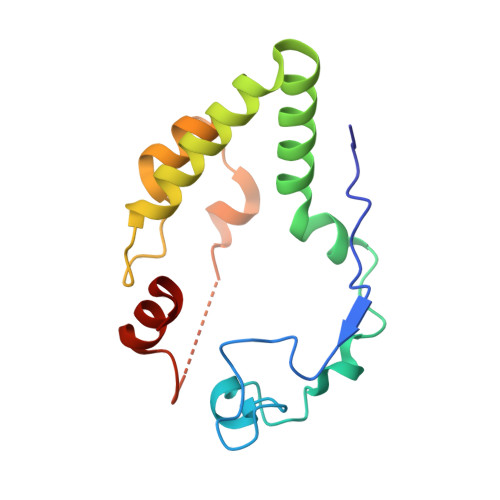
Mechanism of autocatalytic activation during proteasome assembly.
Velez, B., Walsh Jr., R.M., Rawson, S., Razi, A., Adams, L., Perez, E.F., Jiao, F., Blickling, M., Rajakumar, T., Fung, D., Huang, L., Hanna, J.(2024) Nat Struct Mol Biol 31: 1167-1175
- PubMed: 38600323
- DOI: https://doi.org/10.1038/s41594-024-01262-1
- Primary Citation of Related Structures:
8U6Y, 8U7U - PubMed Abstract:
Many large molecular machines are too elaborate to assemble spontaneously and are built through ordered pathways orchestrated by dedicated chaperones. During assembly of the core particle (CP) of the proteasome, where protein degradation occurs, its six active sites are simultaneously activated via cleavage of N-terminal propeptides. Such activation is autocatalytic and coupled to fusion of two half-CP intermediates, which protects cells by preventing activation until enclosure of the active sites within the CP interior. Here we uncover key mechanistic aspects of autocatalytic activation, which proceeds through alignment of the β5 and β2 catalytic triad residues, respectively, with these triads being misaligned before fusion. This mechanism contrasts with most other zymogens, in which catalytic centers are preformed. Our data also clarify the mechanism by which individual subunits can be added in a precise, temporally ordered manner. This work informs two decades-old mysteries in the proteasome field, with broader implications for protease biology and multisubunit complex assembly.
- Department of Pathology, Harvard Medical School and Brigham and Women's Hospital, Boston, MA, USA.
Organizational Affiliation: