Tamsulosin inhibits the release of Chloride ion through wedging into a novel allosteric site of TMEM16A
Li, H.L., Li, S.L., Li, S.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Anoctamin-1 | 960 | Mus musculus | Mutation(s): 0 Gene Names: Ano1, Tmem16a Membrane Entity: Yes | 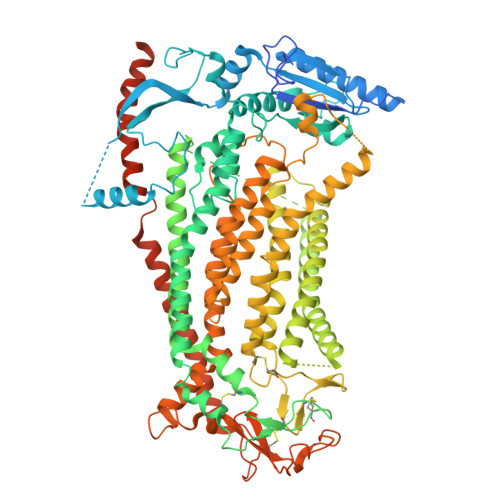 | |
UniProt | |||||
Find proteins for Q8BHY3 (Mus musculus) Explore Q8BHY3 Go to UniProtKB: Q8BHY3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8BHY3 | ||||
Glycosylation | |||||
| Glycosylation Sites: 4 | Go to GlyGen: Q8BHY3-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| JGX (Subject of Investigation/LOI) Query on JGX | GA [auth A], PB [auth B] | Tamsulosin C20 H28 N2 O5 S DRHKJLXJIQTDTD-OAHLLOKOSA-N |  | ||
| NAG Query on NAG | C [auth A] D [auth A] E [auth A] F [auth A] KA [auth B] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| C14 Query on C14 | AA [auth A] AB [auth B] BA [auth A] BB [auth B] CA [auth A] | TETRADECANE C14 H30 BGHCVCJVXZWKCC-UHFFFAOYSA-N |  | ||
| HEX Query on HEX | HA [auth A], JA [auth B] | HEXANE C6 H14 VLKZOEOYAKHREP-UHFFFAOYSA-N |  | ||
| CA Query on CA | EA [auth A], FA [auth A], NB [auth B], OB [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | 1.20.1_4487: |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | -- |