Crystal structure of L-ribulose 3-epimerase in complex with D-allulose
Watanabe, M., Nakamichi, Y., Mine, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ketose 3-epimerase | 297 | Arthrobacter globiformis | Mutation(s): 0 Gene Names: DAE EC: 5.1.3 | 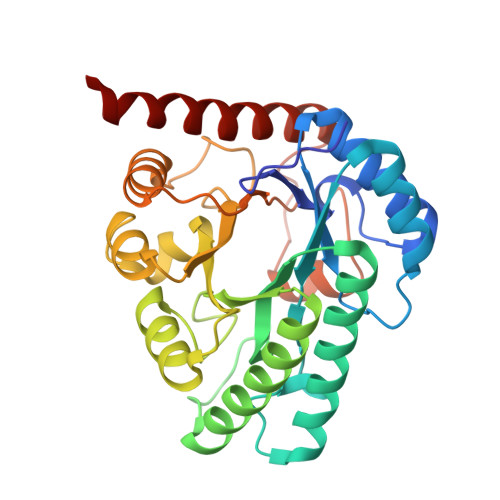 | |
UniProt | |||||
Find proteins for A0A1L7NQ96 (Arthrobacter globiformis) Explore A0A1L7NQ96 Go to UniProtKB: A0A1L7NQ96 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1L7NQ96 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FUD (Subject of Investigation/LOI) Query on FUD | BB [auth K] CA [auth B] FA [auth C] FB [auth L] IB [auth M] | D-fructose C6 H12 O6 BJHIKXHVCXFQLS-UYFOZJQFSA-N |  | ||
| PSJ (Subject of Investigation/LOI) Query on PSJ | AB [auth K] BA [auth B] EA [auth C] EB [auth L] HB [auth M] | D-psicose C6 H12 O6 BJHIKXHVCXFQLS-PUFIMZNGSA-N |  | ||
| PEG (Subject of Investigation/LOI) Query on PEG | IA [auth C] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| MG (Subject of Investigation/LOI) Query on MG | CB [auth K] DA [auth B] GA [auth C] GB [auth L] JB [auth M] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA (Subject of Investigation/LOI) Query on NA | AA [auth A] DB [auth K] HA [auth C] KB [auth N] PA [auth G] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 130.425 | α = 90 |
| b = 135.131 | β = 90.06 |
| c = 151.131 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |