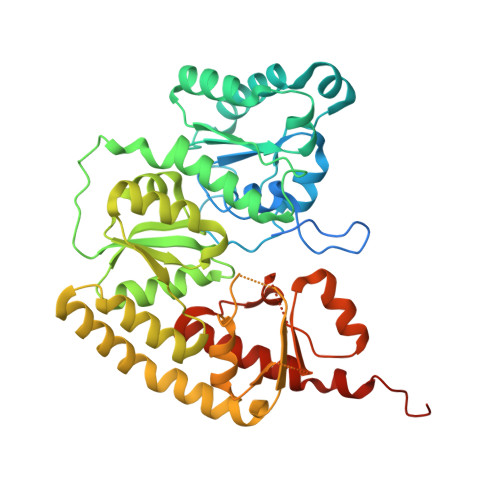
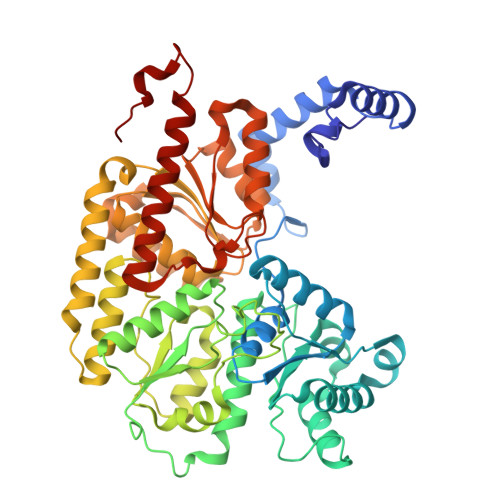
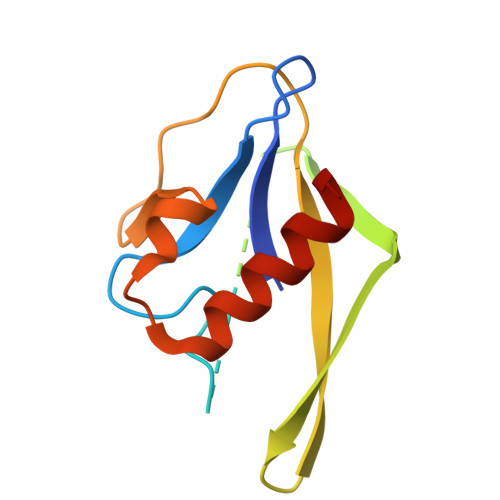
Structural evolution of nitrogenase states under alkaline turnover.
Warmack, R.A., Rees, D.C.(2024) Nat Commun 15: 10472-10472
- PubMed: 39622820
- DOI: https://doi.org/10.1038/s41467-024-54713-0
- Primary Citation of Related Structures:
9CJB, 9CJC, 9CJD, 9CJE, 9CJF - PubMed Abstract:
Biological nitrogen fixation, performed by the enzyme nitrogenase, supplies nearly 50% of the bioavailable nitrogen pool on Earth, yet the structural nature of the enzyme intermediates involved in this cycle remains ambiguous. Here we present four high resolution cryoEM structures of the nitrogenase MoFe-protein, sampled along a time course of alkaline reaction mixtures under an acetylene atmosphere. This series of structures reveals a sequence of salient changes including perturbations to the inorganic framework of the FeMo-cofactor; depletion of the homocitrate moiety; diminished density around the S2B belt sulfur of the FeMo-cofactor; rearrangements of cluster-adjacent side chains; and the asymmetric displacement of the FeMo-cofactor. We further demonstrate that the nitrogenase associated factor T protein can recognize and bind an alkaline inactivated MoFe-protein in vitro. These time-resolved structures provide experimental support for the displacement of S2B and distortions of the FeMo-cofactor at the E 0 -E 3 intermediates of the substrate reduction mechanism, prior to nitrogen binding, highlighting cluster rearrangements potentially relevant to nitrogen fixation by biological and synthetic clusters.
- Division of Chemistry and Chemical Engineering 147-75 California Institute of Technology, Pasadena, CA, USA. rwarmack@caltech.edu.
Organizational Affiliation: