Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
Li, Z., Harikrishnan, L.S., Xu, G., Samanta, D., Clemente, J.C., Leng, L., Tu, W., Yang, L., Huang, L., Wang, M., Wang, S., Deng, Q., Behshad, E., Nagilla, R., Orth, P., Rice, C., Strickland, C., Mohammad, H.P., Priestley, E.S., Sui, Z.(2024) J Med Chem
- PubMed: 39570797
- DOI: https://doi.org/10.1021/acs.jmedchem.4c01878
- Primary Citation of Related Structures:
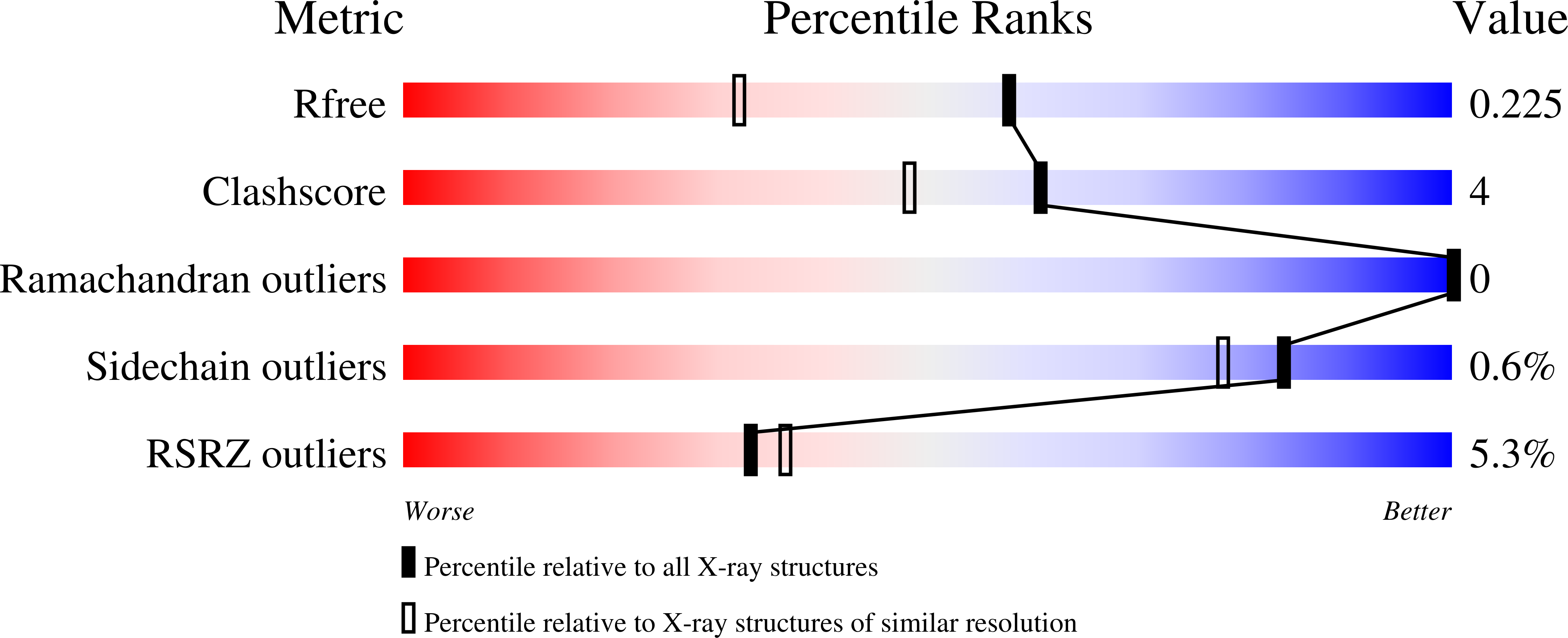
9E1K, 9E30, 9E31 - PubMed Abstract:
We describe the identification of selective SMARCA2, VHL-based heterobifunctional degraders. Structurally novel indolo[1,2- a ]quinazolin-5(7 H )-one SMARCA bromodomain binders were optimized and then converted to SMARCA2 degraders by linking them to well-defined VHL ligands. Our exploration led to the discovery of potent and selective degraders of SMARCA2 over the SMARCA4 paralog, leading to potent and selective growth inhibition of SMARCA4 mutant versus wild type cell lines. We further highlight the optimization of the pharmacokinetic profile of a subset of compounds leading to potent and selective degradation of SMARCA2 in the xenograft model. These compounds provide valuable tools with desirable properties for continued exploration of the biology defining the susceptibility of SMARCA4 mutant cancers to selective loss of SMARCA2.
Organizational Affiliation:
SK Life Science Labs, 2500 Renaissance Blvd, King of Prussia, Pennsylvania 19406, United States.