An Rhs effector uses distinct target cell functions to intoxicate bacterial and fungal competitors
Avelar, G.M., Pankov, G., Sarapa, T., Quinn, J., Hunter, W.N., Rickman, C., Coulthurst, S.J.(2025) bioRxiv
Experimental Data Snapshot
Starting Models: experimental, in silico
View more details
(2025) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Elongation factor Tu 2 | A [auth C], D [auth F] | 394 | Escherichia coli | Mutation(s): 0 Gene Names: tufB, b3980, JW3943 EC: 3.6.5.3 | 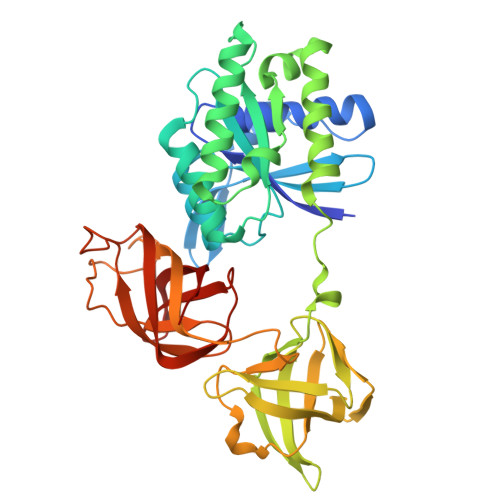 |
UniProt | |||||
Find proteins for P0CE48 (Escherichia coli (strain K12)) Explore P0CE48 Go to UniProtKB: P0CE48 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0CE48 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Immunity protein RhsI2 | 162 | Serratia marcescens | Mutation(s): 0 Gene Names: FBF90_11815 | 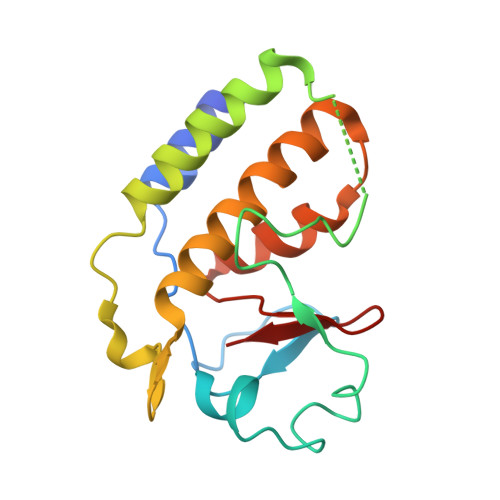 | |
UniProt | |||||
Find proteins for A0ABF7SXG6 (Serratia marcescens) Explore A0ABF7SXG6 Go to UniProtKB: A0ABF7SXG6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0ABF7SXG6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Rhs-family protein | C [auth A], F [auth D] | 162 | Serratia marcescens | Mutation(s): 0 Gene Names: SMDB11_1610 | 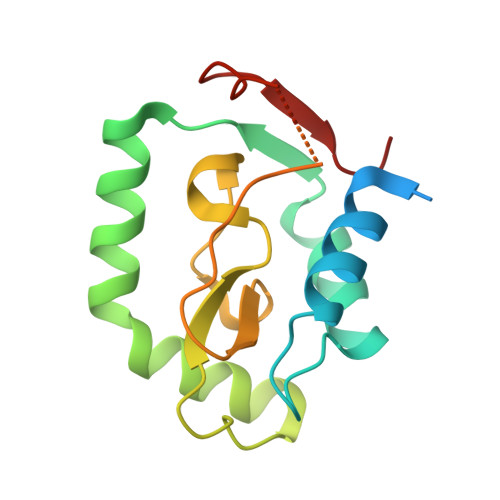 |
UniProt | |||||
Find proteins for A0ABC9II69 (Serratia marcescens subsp. marcescens Db11) Explore A0ABC9II69 Go to UniProtKB: A0ABC9II69 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0ABC9II69 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GDP (Subject of Investigation/LOI) Query on GDP | G [auth C], K [auth F] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| IMD Query on IMD | J [auth C], N [auth F] | IMIDAZOLE C3 H5 N2 RAXXELZNTBOGNW-UHFFFAOYSA-O |  | ||
| EDO Query on EDO | H [auth C], L [auth F] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| MG Query on MG | I [auth C], M [auth F] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 61.598 | α = 90 |
| b = 110.569 | β = 102.392 |
| c = 113.863 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| DIALS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Wellcome Trust | United Kingdom | 215599/Z/19/Z |
| Wellcome Trust | United Kingdom | 220321/Z/20/Z |