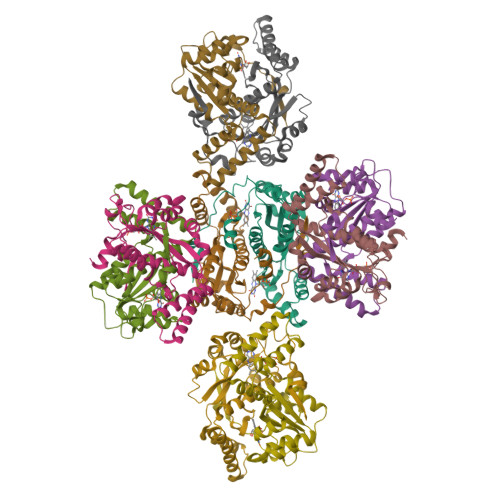
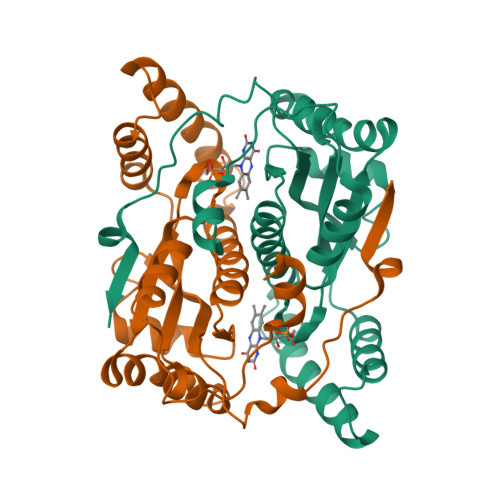
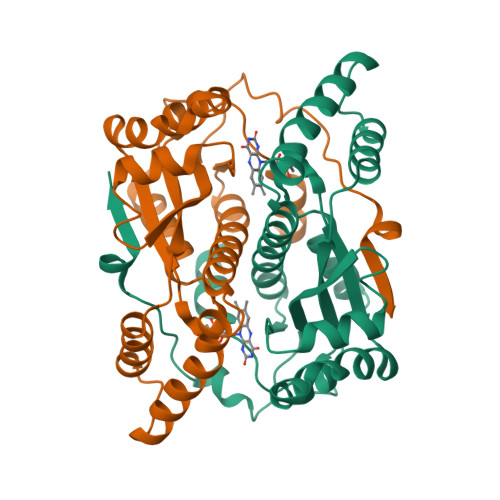
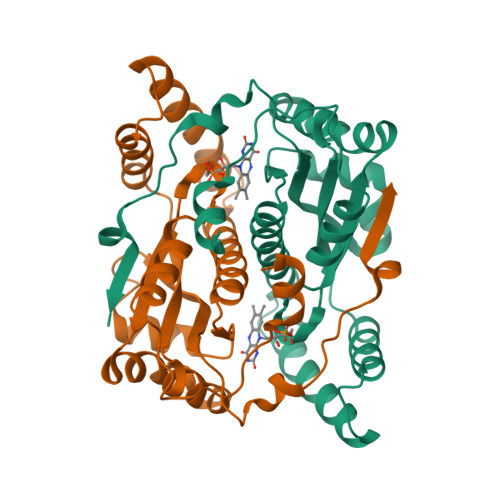
Altering the regioselectivity of a nitroreductase in the synthesis of arylhydroxylamines by structure-based engineering.
Bai, J., Zhou, Y., Chen, Q., Yang, Q., Yang, J.(2015) Chembiochem 16: 1219-1225
- PubMed: 25917861
- DOI: https://doi.org/10.1002/cbic.201500070
- Primary Citation of Related Structures:
3X21 - PubMed Abstract:
Nitroreductases have great potential for the highly efficient reduction of aryl nitro compounds to arylhydroxylamines. However, regioselective reduction of the desired nitro group in polynitroarenes is still a challenge. Here, we describe the structure-based engineering of Escherichia coli nitroreductase NfsB to alter its regioselectivity, in order to achieve reduction of a target nitro group. When 2,4-dinitrotoluene was used as the substrate, the wild-type enzyme regioselectively reduced the 4-NO2 group, but the T41L/N71S/F124W mutant primarily reduced the 2-NO2 group, without loss of activity. The crystal structure of T41L/N71S/F124W and docking experiments indicated that the regioselectivity change (from 4-NO2 to 2-NO2 ) might result from the increased hydrophobicity of residues 41 and 124 (proximal to FMN) and conformational changes in residues 70 and 124.
Organizational Affiliation:
School of Life Science and Biotechnology, Dalian University of Technology, No. 2 Linggong Road, Dalian-116023 (China).