Two high-resolution crystal structures of the recombinant N-lobe of human transferrin reveal a structural change implicated in iron release.
MacGillivray, R.T., Moore, S.A., Chen, J., Anderson, B.F., Baker, H., Luo, Y., Bewley, M., Smith, C.A., Murphy, M.E., Wang, Y., Mason, A.B., Woodworth, R.C., Brayer, G.D., Baker, E.N.(1998) Biochemistry 37: 7919-7928
- PubMed: 9609685
- DOI: https://doi.org/10.1021/bi980355j
- Primary Citation of Related Structures:
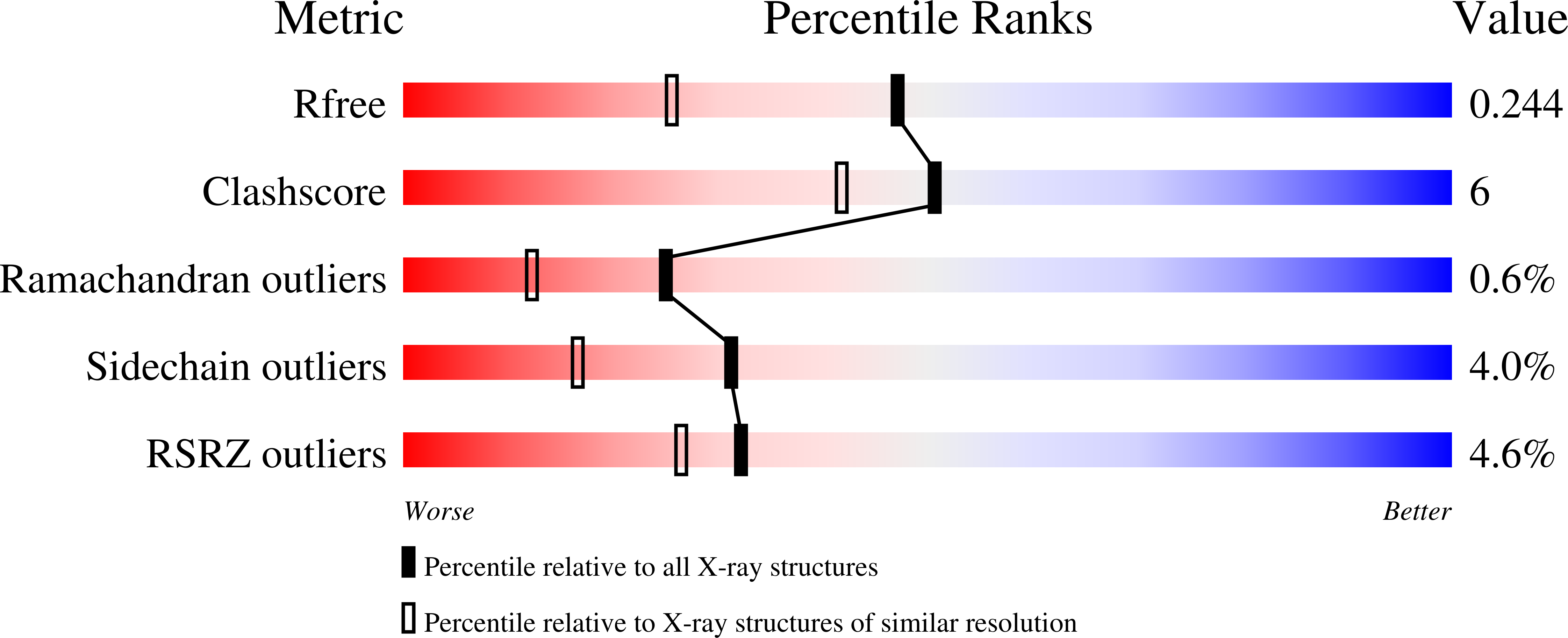
1A8E, 1A8F - PubMed Abstract:
The N-lobe of human serum transferrin (hTF/2N) has been expressed in baby hamster kidney cells and crystallized in both orthorhombic (P212121) and tetragonal (P41212) space groups. Both crystal forms diffract to high resolution (1.6 and 1.8 A, respectively) and have been solved by molecular replacement. Subsequent refinement resulted in final models for the structure of hTF/2N that had crystallographic R-factors of 18.1 and 19.7% for the two crystal forms, respectively; these models represent the highest-resolution transferrin structures determined to date. The hTF/2N polypeptide has a folding pattern similar to those of other transferrins, including the presence of a deep cleft that contains the metal-binding site. In contrast to other transferrins, both crystal forms of hTF/2N display disorder at the iron-binding site; model building suggests that this disorder consists of alternative conformations of the synergistically bound carbonate anion, the side chain for Arg-124, and several solvent molecules. Subsequent refinement revealed that conformation A has an occupancy of 0.63-0. 65 and corresponds to the structure of the iron-binding site found in other transferrins. The alternative conformation B has an occupancy of 0.35-0.37; in this structure, the carbonate has rotated 30 degrees relative to the iron and the side chain for Arg-124 has moved to accommodate the new carbonate position. Several water molecules appear to stabilize the carbonate anion in the two conformations. These structures are consistent with the protonation of the carbonate and resulting partial removal of the anion from the metal; these events would occur prior to cleft opening and metal release.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of British Columbia, Vancouver, Canada. macg@unixg.ubc.ca