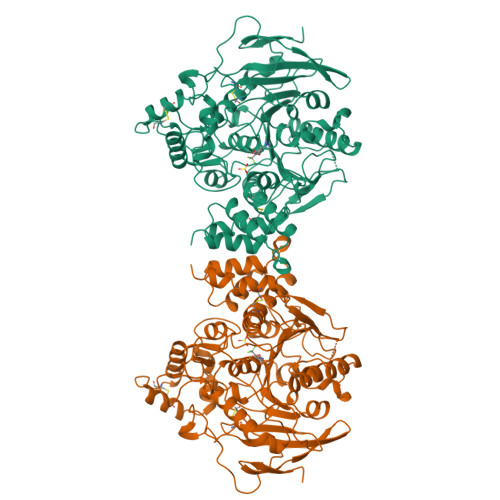
The X-ray structure of a transition state analog complex reveals the molecular origins of the catalytic power and substrate specificity of acetylcholinesterase.
Harel, M., Quinn, D.M., Nair, H.K., Silman, I., Sussman, J.L.(1996) J Am Chem Soc 118: 2340-2346