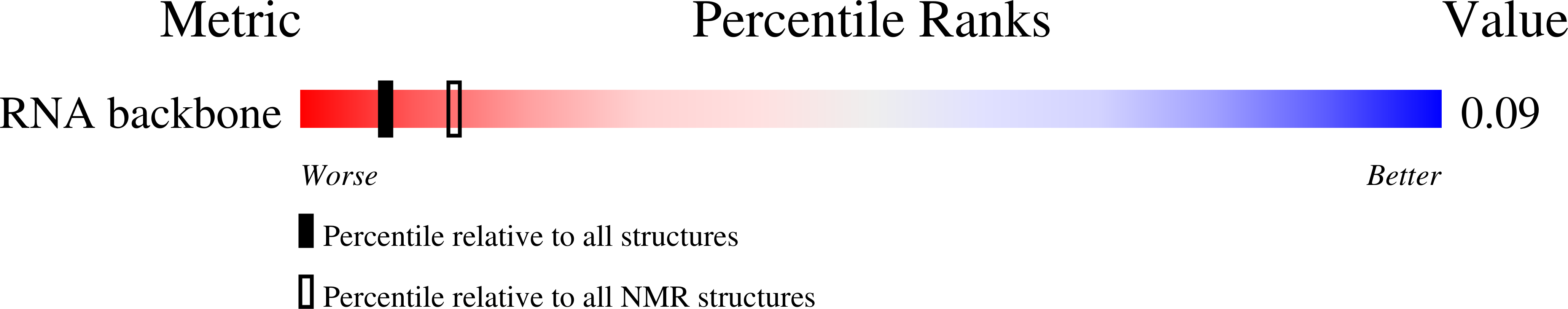Solution structure of an RNA duplex including a C-U base pair.
Tanaka, Y., Kojima, C., Yamazaki, T., Kodama, T.S., Yasuno, K., Miyashita, S., Ono, A., Ono, A., Kainosho, M., Kyogoku, Y.(2000) Biochemistry 39: 7074-7080
- PubMed: 10852704
- DOI: https://doi.org/10.1021/bi000018m
- Primary Citation of Related Structures:
1C4L - PubMed Abstract:
The formation of the C-U base pair in a duplex was observed in solution by means of the temperature profile of (15)N chemical shifts, and the precise geometry of the C-U base pair was also determined by NOE-based structure calculation. From the solution structure of the RNA oligomer, r[CGACUCAGG].r[CCUGCGUCG], it was found that a single C-U mismatch preferred being stacked in the duplex rather than being flipped-out even in solution. Moreover, it adopts an irregular geometry, where the amino nitrogen (N4) of the cytidine and keto-oxygen (O4) of the uridine are within hydrogen-bonding distance, as seen in crystals. To further prove the presence of a hydrogen bond in the C-U pair, we employed a point-labeled cytidine at the exocyclic amino nitrogen of the cytidine in the C-U pair. The temperature profile of its (15)N chemical shift showed a sigmoidal transition curve, indicating the presence of a hydrogen bond in the C-U pair in the duplex.
Organizational Affiliation:
Institute for Protein Research, Osaka University, Japan.