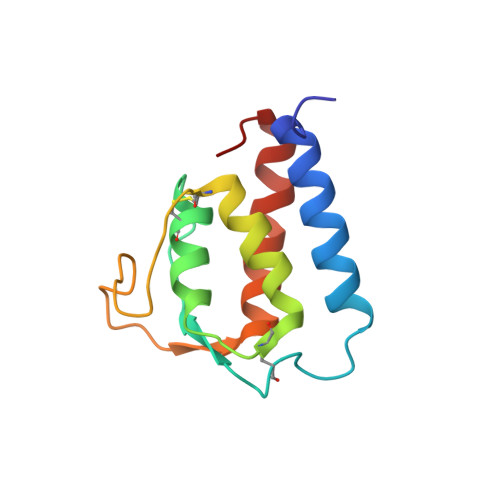
Solution structure of human IL-13 and implication for receptor binding.
Moy, F.J., Diblasio, E., Wilhelm, J., Powers, R.(2001) J Mol Biology 310: 219-230
- PubMed: 11419948
- DOI: https://doi.org/10.1006/jmbi.2001.4764
- Primary Citation of Related Structures:
1IJZ, 1IK0 - PubMed Abstract:
Interleukin-13 has been implicated as a key factor in asthma, allergy, atopy and inflammatory response, establishing the protein as a valuable therapeutic target. The high-resolution solution structure of human IL-13 has been determined by multidimensional NMR. The resulting structure is consistent with previous short-chain left-handed four-helix bundles, where a significant similarity in the folding topology between IL-13 and IL-4 was observed. IL-13 shares a significant overlap in biological function with IL-4, a result of the common alpha chain component (IL-4Ralpha) in their respective receptors. Based on the available structural and mutational data, an IL-13/IL-4Ralpha model and a sequential mechanism for forming the signaling heterodimer is proposed for IL-13.
- Department of Biological Chemistry,Wyeth Research, Cambridge, MA 02140, USA.
Organizational Affiliation: