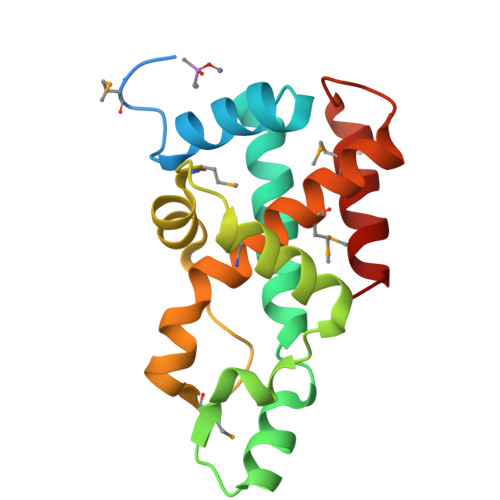
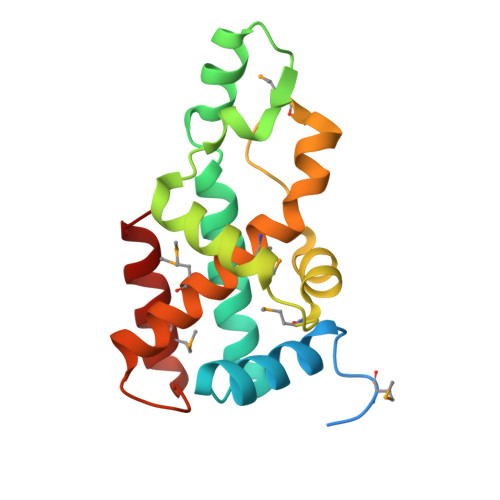
The crystal structure of Aq_328 from the hyperthermophilic bacteria Aquifex aeolicus shows an ancestral histone fold.
Qiu, Y., Tereshko, V., Kim, Y., Zhang, R., Collart, F., Yousef, M., Kossiakoff, A., Joachimiak, A.(2006) Proteins 62: 8-16
- PubMed: 16287087
- DOI: https://doi.org/10.1002/prot.20590
- Primary Citation of Related Structures:
1R4V - PubMed Abstract:
The structure of Aq_328, an uncharacterized protein from hyperthermophilic bacteria Aquifex aeolicus, has been determined to 1.9 A by using multi-wavelength anomalous diffraction (MAD) phasing. Although the amino acid sequence analysis shows that Aq_328 has no significant similarity to proteins with a known structure and function, the structure comparison by using the Dali server reveals that it: (1) assumes a histone-like fold, and (2) is similar to an ancestral nuclear histone protein (PDB code 1F1E) with z-score 8.1 and RMSD 3.6 A over 124 residues. A sedimentation equilibrium experiment indicates that Aq_328 is a monomer in solution, with an average sedimentation coefficient of 2.4 and an apparent molecular weight of about 20 kDa. The overall architecture of Aq_328 consists of two noncanonical histone domains in tandem repeat within a single chain, and is similar to eukaryotic heterodimer (H2A/H2B and H3/H4) and an archaeal histone heterodimer (HMfA/HMfB). The sequence comparisons between the two histone domains of Aq_328 and six eukaryotic/archaeal histones demonstrate that most of the conserved residues that underlie the Aq_328 architecture are used to build and stabilize the two cross-shaped antiparallel histone domains. The high percentage of salt bridges in the structure could be a factor in the protein's thermostability. The structural similarities to other histone-like proteins, molecular properties, and potential function of Aq_328 are discussed in this paper.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, Illinois 60637, USA.