Solution Structure and Antibody Binding Studies of the Envelope Protein Domain III from the New York Strain of West Nile Virus
Volk, D.E., Beasley, D.W., Kallick, D.A., Holbrook, M.R., Barrett, A.D., Gorenstein, D.G.(2004) J Biological Chem 279: 38755-38761
- PubMed: 15190071
- DOI: https://doi.org/10.1074/jbc.M402385200
- Primary Citation of Related Structures:
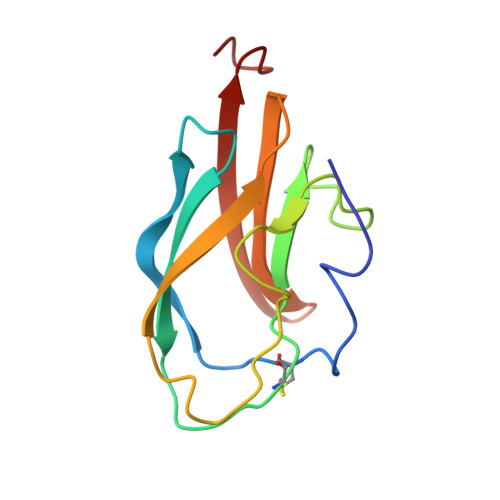
1S6N - PubMed Abstract:
The solution structure of domain III from the New York West Nile virus strain 385-99 (WN-rED3) has been determined by NMR methods. The West Nile domain III structure is a beta-barrel structure formed from seven anti-parallel beta-strands in two beta-sheets. One anti-parallel beta-sheet consists of beta-strands beta1 (Phe(299)-Asp(307)), beta2 (Val(313)-Tyr(319)), beta4 (Arg(354)-Leu(355)), and beta5 (Lys(370)-Glu(376)) arranged so that beta2 is flanked on either side by beta1 and beta5. The short beta4 flanks the end of the remaining side of beta5. The remaining anti-parallel beta-sheet is formed from strands beta3 (Ile(340)-Val(343)), beta6 (Gly(380)-Arg(388)), and beta7 (Gln(391)-Lys(399)) arranged with beta6 at the center. Residues implicated in antigenic differences between different West Nile virus strains (and other flaviviruses) and neutralization are located on the outer surface of the protein. Characterization of the binding of monoclonal antibodies to WN-rED3 mutants, which were identified through neutralization escape experiments, indicate that antibody neutralization directly correlates with binding affinities. These studies provide an insight into theoretical virus-receptor interaction points, structure of immunogenic determinants, and potential targets for antiviral agents against West Nile virus and highlight differences between West Nile virus and other flavivirus structures that may represent critical determinants of virulence.
- Sealy Center for Structural Biology, University of Texas Medical Branch, Galveston, Texas 77555-1147, USA.
Organizational Affiliation: