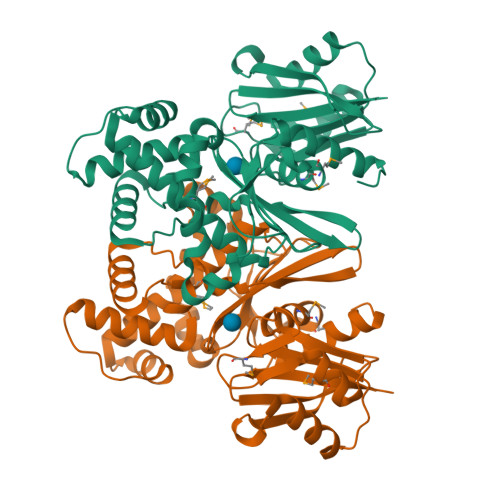
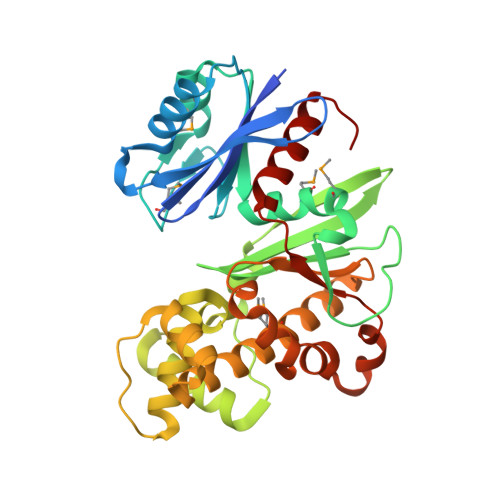
Crystal structures of Escherichia coli ATP-dependent glucokinase and its complex with glucose
Lunin, V.V., Li, Y., Schrag, J.D., Iannuzzi, P., Cygler, M., Matte, A.(2004) J Bacteriol 186: 6915-6927
- PubMed: 15466045
- DOI: https://doi.org/10.1128/JB.186.20.6915-6927.2004
- Primary Citation of Related Structures:
1Q18, 1SZ2 - PubMed Abstract:
Intracellular glucose in Escherichia coli cells imported by phosphoenolpyruvate-dependent phosphotransferase system-independent uptake is phosphorylated by glucokinase by using ATP to yield glucose-6-phosphate. Glucokinases (EC 2.7.1.2) are functionally distinct from hexokinases (EC 2.7.1.1) with respect to their narrow specificity for glucose as a substrate. While structural information is available for ADP-dependent glucokinases from Archaea, no structural information exists for the large sequence family of eubacterial ATP-dependent glucokinases. Here we report the first structure determination of a microbial ATP-dependent glucokinase, that from E. coli O157:H7. The crystal structure of E. coli glucokinase has been determined to a 2.3-A resolution (apo form) and refined to final Rwork/Rfree factors of 0.200/0.271 and to 2.2-A resolution (glucose complex) with final Rwork/Rfree factors of 0.193/0.265. E. coli GlK is a homodimer of 321 amino acid residues. Each monomer folds into two domains, a small alpha/beta domain (residues 2 to 110 and 301 to 321) and a larger alpha+beta domain (residues 111 to 300). The active site is situated in a deep cleft between the two domains. E. coli GlK is structurally similar to Saccharomyces cerevisiae hexokinase and human brain hexokinase I but is distinct from the ADP-dependent GlKs. Bound glucose forms hydrogen bonds with the residues Asn99, Asp100, Glu157, His160, and Glu187, all of which, except His160, are structurally conserved in human hexokinase 1. Glucose binding results in a closure of the small domains, with a maximal Calpha shift of approximately 10 A. A catalytic mechanism is proposed that is consistent with Asp100 functioning as the general base, abstracting a proton from the O6 hydroxyl of glucose, followed by nucleophilic attack at the gamma-phosphoryl group of ATP, yielding glucose-6-phosphate as the product.
Organizational Affiliation:
Biotechnology Research Institute, NRCC, 6100 Royalmount Ave., Montreal, Quebec H4P 2R2 Canada.