The complex of Bacillus pasteurii urease with beta-mercaptoethanol from X-ray data at 1.65-A resolution
Benini, S., Rypniewski, W.R., Wilson, K.S., Ciurli, S., Mangani, S.(1998) J Biol Inorg Chem 3: 268-273
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(1998) J Biol Inorg Chem 3: 268-273
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UREASE | 100 | Sporosarcina pasteurii | Mutation(s): 0 EC: 3.5.1.5 | 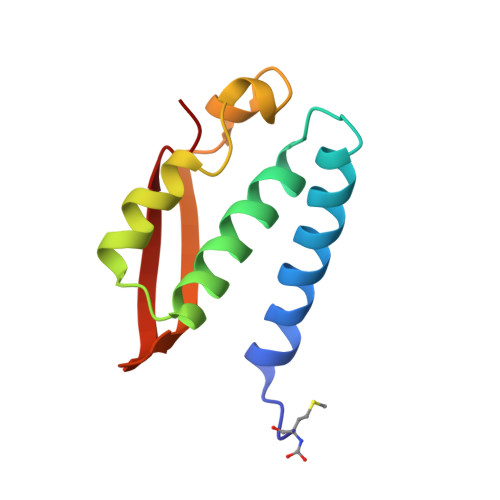 | |
UniProt | |||||
Find proteins for P41022 (Sporosarcina pasteurii) Explore P41022 Go to UniProtKB: P41022 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P41022 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UREASE | 122 | Sporosarcina pasteurii | Mutation(s): 0 EC: 3.5.1.5 | 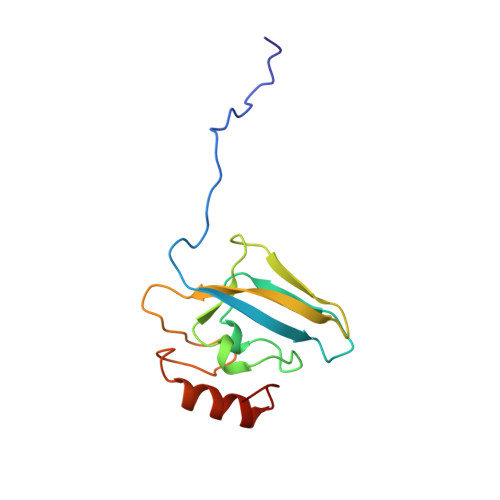 | |
UniProt | |||||
Find proteins for P41021 (Sporosarcina pasteurii) Explore P41021 Go to UniProtKB: P41021 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P41021 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| UREASE | 570 | Sporosarcina pasteurii | Mutation(s): 8 EC: 3.5.1.5 | 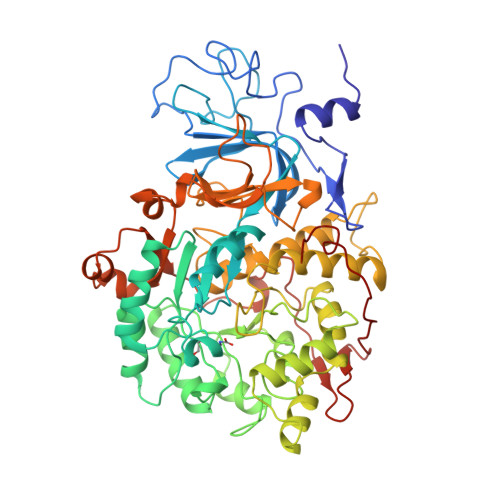 | |
UniProt | |||||
Find proteins for P41020 (Sporosarcina pasteurii) Explore P41020 Go to UniProtKB: P41020 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P41020 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BME Query on BME | F [auth C], G [auth C] | BETA-MERCAPTOETHANOL C2 H6 O S DGVVWUTYPXICAM-UHFFFAOYSA-N |  | ||
| NI Query on NI | D [auth C], E [auth C] | NICKEL (II) ION Ni VEQPNABPJHWNSG-UHFFFAOYSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CXM Query on CXM | A | L-PEPTIDE LINKING | C6 H11 N O4 S |  | MET |
| KCX Query on KCX | C | L-PEPTIDE LINKING | C7 H14 N2 O4 |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 131.343 | α = 90 |
| b = 131.343 | β = 90 |
| c = 190.013 | γ = 120 |
| Software Name | Purpose |
|---|---|
| AMoRE | phasing |
| REFMAC | refinement |
| DENZO | data reduction |
| SCALEPACK | data scaling |