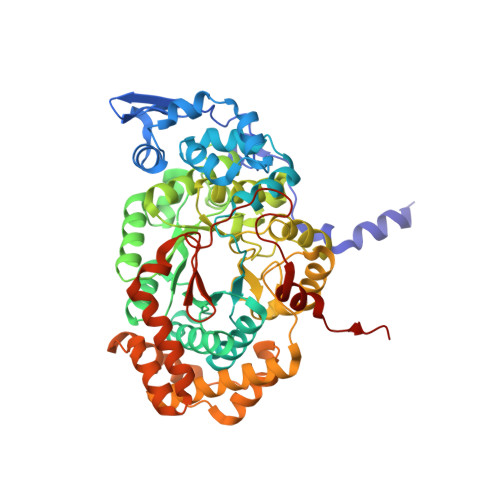
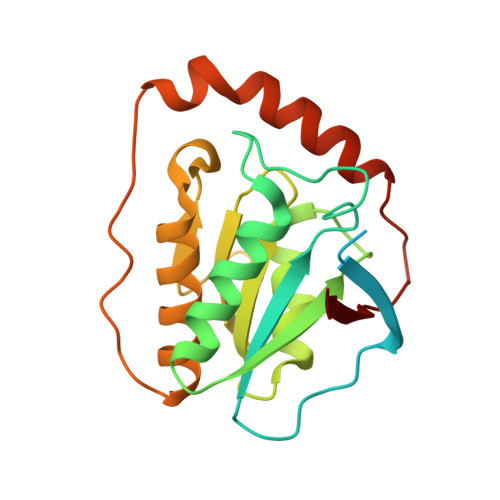
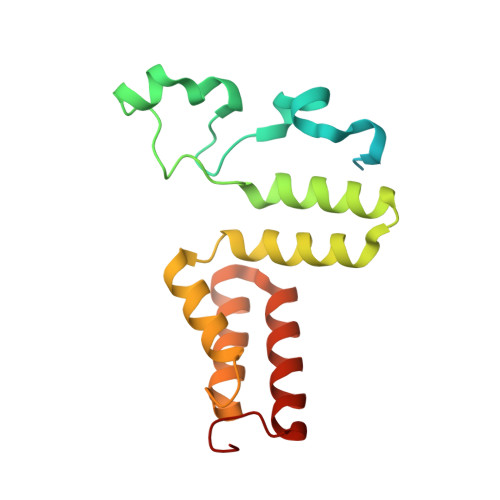
Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
Shibata, N., Nakanishi, Y., Fukuoka, M., Yamanishi, M., Yasuoka, N., Toraya, T.(2003) J Biological Chem 278: 22717-22725
- PubMed: 12684496
- DOI: https://doi.org/10.1074/jbc.M301513200
- Primary Citation of Related Structures:
1UC4, 1UC5 - PubMed Abstract:
Adenosylcobalamin-dependent diol dehydratase of Klebsiella oxytoca is apparently not stereospecific and catalyzes the conversion of both (R)- and (S)-1,2-propanediol to propionaldehyde. To explain this unusual property of the enzyme, we analyzed the crystal structures of diol dehydratase in complexes with cyanocobalamin and (R)- or (S)-1,2-propanediol. (R)- and (S)-isomers are bound in a symmetrical manner, although the hydrogen-bonding interactions between the substrate and the active-site residues are the same. From the position of the adenosyl radical in the modeled "distal" conformation, it is reasonable for the radical to abstract the pro-R and pro-S hydrogens from (R)- and (S)-isomers, respectively. The hydroxyl groups in the substrate radicals would migrates from C(2) to C(1) by a suprafacial shift, resulting in the stereochemical inversion at C(1). This causes 60 degrees clockwise and 70 degrees counterclockwise rotations of the C(1)-C(2) bond of the (R)- and (S)-isomers, respectively, if viewed from K+. A modeling study of 1,1-gem-diol intermediates indicated that new radical center C(2) becomes close to the methyl group of 5'-deoxyadenosine. Thus, the hydrogen back-abstraction (recombination) from 5'-deoxyadenosine by the product radical is structurally feasible. It was also predictable that the substitution of the migrating hydroxyl group by a hydrogen atom from 5'-deoxyadenosine takes place with the inversion of the configuration at C(2) of the substrate. Stereospecific dehydration of the 1,1-gem-diol intermediates can also be rationalized by assuming that Asp-alpha335 and Glu-alpha170 function as base catalysts in the dehydration of the (R)- and (S)-isomers, respectively. The structure-based mechanism and stereochemical courses of the reaction are proposed.
- Department of Life Science, Graduate School of Science, Himeji Institute of Technology, 3-2-1 Kouto, Kamigori, Ako-gun, Hyogo 678-1297, Japan.
Organizational Affiliation: