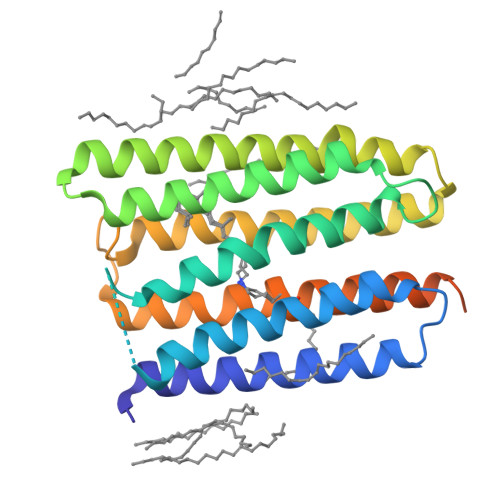
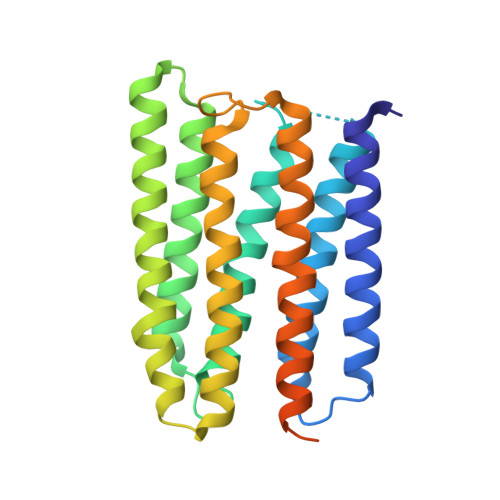
Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A.
Vogeley, L., Sineshchekov, O.A., Trivedi, V.D., Sasaki, J., Spudich, J.L., Luecke, H.(2004) Science 306: 1390-1393
- PubMed: 15459346
- DOI: https://doi.org/10.1126/science.1103943
- Primary Citation of Related Structures:
1XIO - PubMed Abstract:
Microbial sensory rhodopsins are a family of membrane-embedded photoreceptors in prokaryotic and eukaryotic organisms. Structures of archaeal rhodopsins, which function as light-driven ion pumps or photosensors, have been reported. We present the structure of a eubacterial rhodopsin, which differs from those of previously characterized archaeal rhodopsins in its chromophore and cytoplasmic-side portions. Anabaena sensory rhodopsin exhibits light-induced interconversion between stable 13-cis and all-trans states of the retinylidene protein. The ratio of its cis and trans chromophore forms depends on the wavelength of illumination, thus providing a mechanism for a single protein to signal the color of light, for example, to regulate color-sensitive processes such as chromatic adaptation in photosynthesis. Its cytoplasmic half channel, highly hydrophobic in the archaeal rhodopsins, contains numerous hydrophilic residues networked by water molecules, providing a connection from the photoactive site to the cytoplasmic surface believed to interact with the receptor's soluble 14-kilodalton transducer.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697, USA.