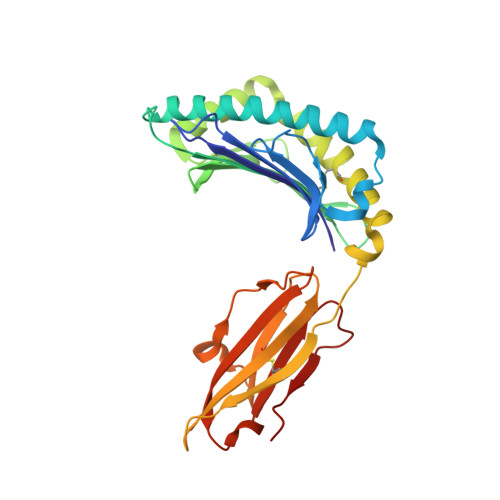
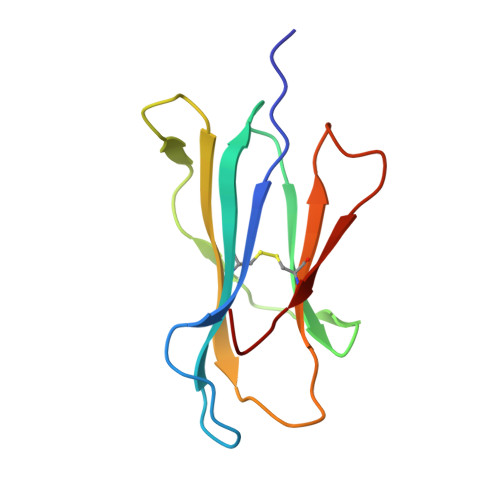
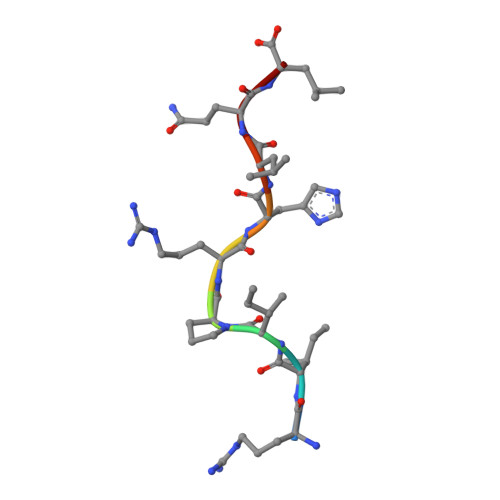
Crystal structure of HLA-G: A nonclassical MHC class I molecule expressed at the fetal-maternal interface
Clements, C.S., Kjer-Nielsen, L., Kostenko, L., Hoare, H.L., Dunstone, M.A., Moses, E., Freed, K., Brooks, A.G., Rossjohn, J., McCluskey, J.(2005) Proc Natl Acad Sci U S A 102: 3360-3365
- PubMed: 15718280
- DOI: https://doi.org/10.1073/pnas.0409676102
- Primary Citation of Related Structures:
1YDP - PubMed Abstract:
HLA-G is a nonclassical major histocompatibility complex class I (MHC-I) molecule that is primarily expressed at the fetal-maternal interface, where it is thought to play a role in protecting the fetus from the maternal immune response. HLA-G binds a limited repertoire of peptides and interacts with the inhibitory leukocyte Ig-like receptors LIR-1 and LIR-2 and possibly with certain natural killer cell receptors. To gain further insights into HLA-G function, we determined the 1.9-A structure of a monomeric HLA-G complexed to a natural endogenous peptide ligand from histone H2A (RIIPRHLQL). An extensive network of contacts between the peptide and the antigen-binding cleft reveal a constrained mode of binding reminiscent of the nonclassical HLA-E molecule, thereby providing a structural basis for the limited peptide repertoire of HLA-G. The alpha3 domain of HLA-G, a candidate binding site for the LIR-1 and -2 inhibitory receptors, is structurally distinct from the alpha3 domains of classical MHC-I molecules, providing a rationale for the observed affinity differences for these ligands. The structural data suggest a head-to-tail mode of dimerization, mediated by an intermolecular disulfide bond, that is consistent with the observation of HLA-G dimers on the cell surface.
- Protein Crystallography Unit, Monash Centre for Synchrotron Science, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.
Organizational Affiliation: