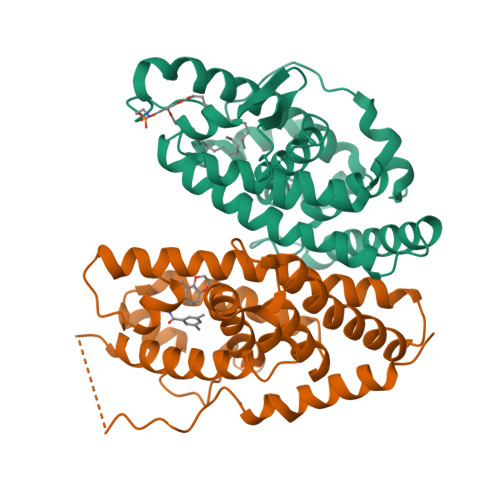
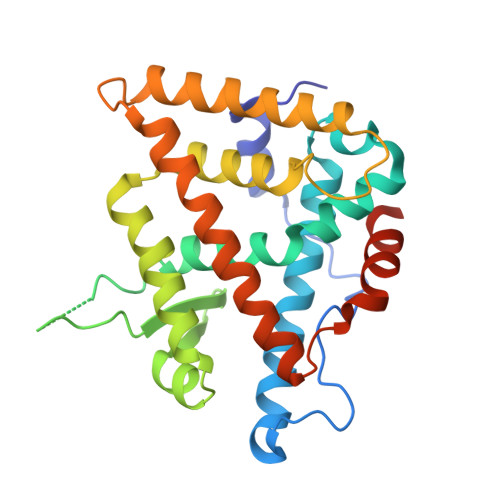
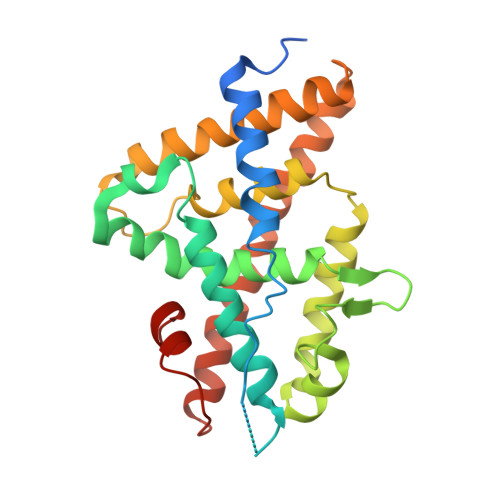
Structural adaptability in the ligand-binding pocket of the ecdysone hormone receptor.
Billas, I.M.L., Iwema, T., Garnier, J.M., Mitschler, A., Rochel, N., Moras, D.(2003) Nature 426: 91-96
- PubMed: 14595375
- DOI: https://doi.org/10.1038/nature02112
- Primary Citation of Related Structures:
1R1K, 1R20 - PubMed Abstract:
The ecdysteroid hormones coordinate the major stages of insect development, notably moulting and metamorphosis, by binding to the ecdysone receptor (EcR); a ligand-inducible nuclear transcription factor. To bind either ligand or DNA, EcR must form a heterodimer with ultraspiracle (USP), the homologue of retinoid-X receptor. Here we report the crystal structures of the ligand-binding domains of the moth Heliothis virescens EcR-USP heterodimer in complex with the ecdysteroid ponasterone A and with a non-steroidal, lepidopteran-specific agonist BYI06830 used in agrochemical pest control. The two structures of EcR-USP emphasize the universality of heterodimerization as a general mechanism common to both vertebrates and invertebrates. Comparison of the EcR structures in complex with steroidal and non-steroidal ligands reveals radically different and only partially overlapping ligand-binding pockets that could not be predicted by molecular modelling and docking studies. These findings offer new perspectives for the design of insect-specific, environmentally safe insecticides. The concept of a ligand-dependent binding pocket in EcR provides an insight into the moulding of nuclear receptors to their ligand, and has potential applications for human nuclear receptors.
Organizational Affiliation:
Département de Biologie et de Génomique Structurales, IGBMC, CNRS/INSERM/Université Louis Pasteur, Parc d'Innovation BP10142, 67404 Illkirch cedex, France.