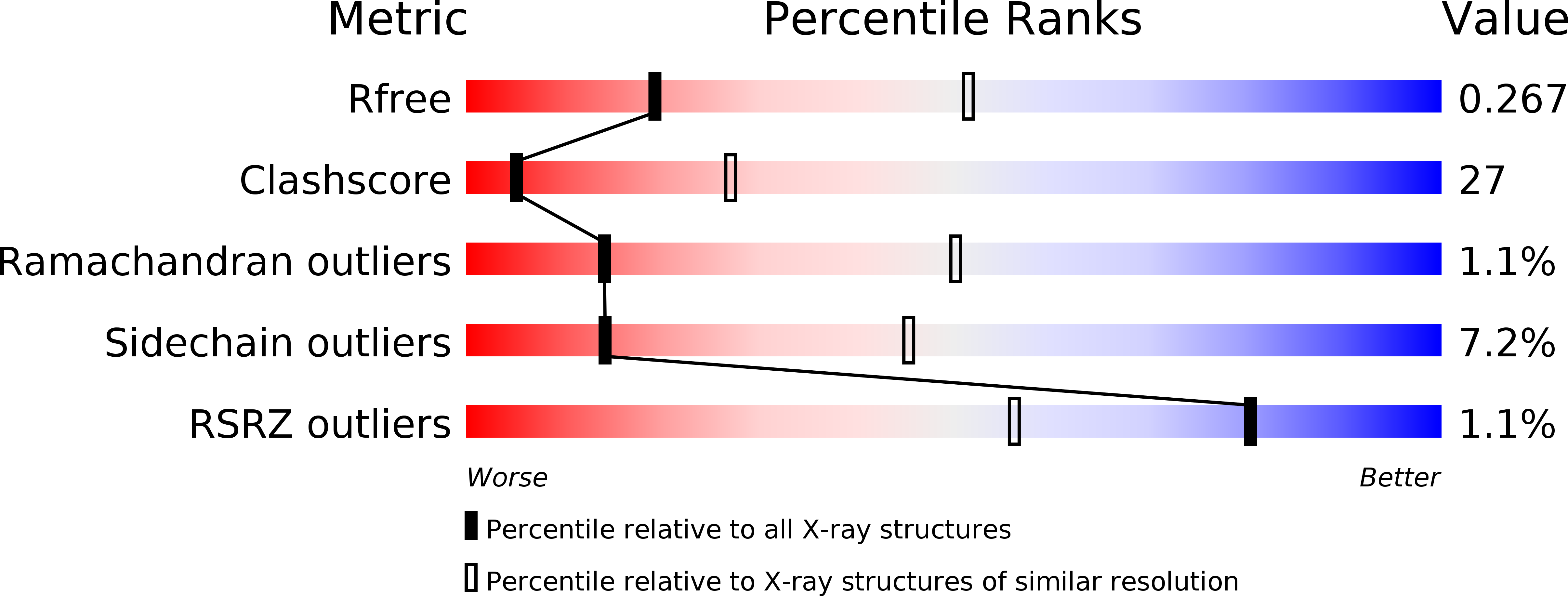
Crystal Structure of Human Guanosine Monophosphate Reductase 2 (GMPR2) in Complex with GMP
Li, J., Wei, Z., Zheng, M., Gu, X., Deng, Y., Qiu, R., Chen, F., Ji, C., Gong, W., Xie, Y., Mao, Y.(2006) J Mol Biol 355: 980-988
- PubMed: 16359702
- DOI: https://doi.org/10.1016/j.jmb.2005.11.047
- Primary Citation of Related Structures:
2A7R - PubMed Abstract:
Guanosine monophosphate reductase (GMPR) catalyzes the irreversible and NADPH-dependent reductive deamination of GMP to IMP, and plays a critical role in re-utilization of free intracellular bases and purine nucleosides. Here, we report the first crystal structure of human GMP reductase 2 (hGMPR2) in complex with GMP at 3.0 A resolution. The protein forms a tetramer composed of subunits adopting the ubiquitous (alpha/beta)8 barrel fold. Interestingly, the substrate GMP is bound to hGMPR2 through interactions with Met269, Ser270, Arg286, Ser288, and Gly290; this makes the conformation of the adjacent flexible binding region (residues 268-289) fixed, much like a door on a hinge. Structure comparison and sequence alignment analyses show that the conformation of the active site loop (residues 179-187) is similar to those of hGMPR1 and inosine monophosphate dehydrogenases (IMPDHs). We propose that Cys186 is the potential active site, and that the conformation of the loop (residues 129-133) suggests a preference for the coenzyme NADPH over NADH. This structure provides important information towards understanding the functions of members of the GMPR family.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Institute of Genetics, School of Life Sciences, Fudan University, Shanghai 200433, People's Republic of China.