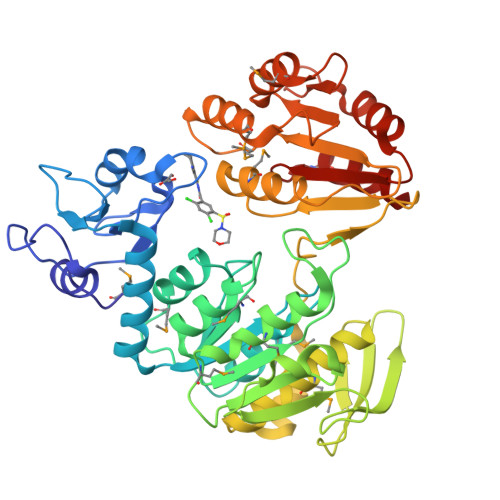
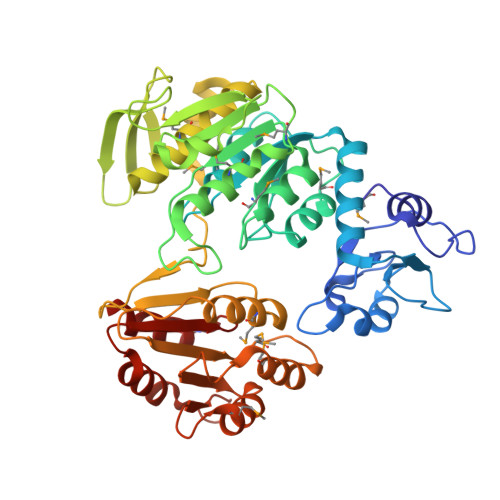
Structure of MurF from Streptococcus pneumoniae co-crystallized with a small molecule inhibitor exhibits interdomain closure
Longenecker, K.L., Stamper, G.F., Hajduk, P.J., Fry, E.H., Jakob, C.G., Harlan, J.E., Edalji, R., Bartley, D.M., Walter, K.A., Solomon, L.R., Holzman, T.F., Gu, Y.G., Lerner, C.G., Beutel, B.A., Stoll, V.S.(2005) Protein Sci 14: 3039-3047
- PubMed: 16322581
- DOI: https://doi.org/10.1110/ps.051604805
- Primary Citation of Related Structures:
2AM1, 2AM2 - PubMed Abstract:
In a broad genomics analysis to find novel protein targets for antibiotic discovery, MurF was identified as an essential gene product for Streptococcus pneumonia that catalyzes a critical reaction in the biosynthesis of the peptidoglycan in the formation of the cell wall. Lacking close relatives in mammalian biology, MurF presents attractive characteristics as a potential drug target. Initial screening of the Abbott small-molecule compound collection identified several compounds for further validation as pharmaceutical leads. Here we report the integrated efforts of NMR and X-ray crystallography, which reveal the multidomain structure of a MurF-inhibitor complex in a compact conformation that differs dramatically from related structures. The lead molecule is bound in the substrate-binding region and induces domain closure, suggestive of the domain arrangement for the as yet unobserved transition state conformation for MurF enzymes. The results form a basis for directed optimization of the compound lead by structure-based design to explore the suitability of MurF as a pharmaceutical target.
Organizational Affiliation:
Department of Structural Biology, R46Y, Building AP10, 100 Abbott Park Road, Abbott Park, IL 60064, USA. Kenton.Longenecker@Abbott.com