X-ray structures of Lqh-alpha-IT and Lqh-alpha-IT8D9D10V mutant
Kahn, R., Karbat, I., Gurevitz, M., Frolow, F.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| neurotoxin alpha-IT | 65 | Leiurus quinquestriatus | Mutation(s): 0 | 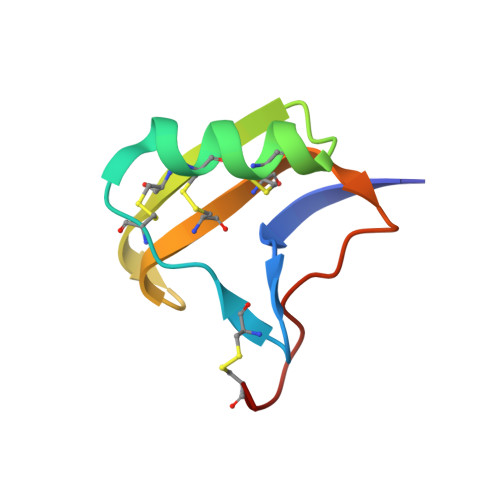 | |
UniProt | |||||
Find proteins for P17728 (Leiurus hebraeus) Explore P17728 Go to UniProtKB: P17728 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P17728 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 8 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| DR0 Query on DR0 | C [auth A] | N-(HYDROXYMETHYL)-N,N-DIMETHYLHEXAN-1-AMINIUM C9 H22 N O HSCKPPPLTPBXED-UHFFFAOYSA-N |  | ||
| DR8 Query on DR8 | B [auth A] | N,N,N-TRIMETHYLHEPTA-1,3,5-TRIYN-1-AMINIUM C10 H12 N AKYJILYZPCHMQA-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | I [auth A], J [auth A], K [auth A], L [auth A], M [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| NO3 Query on NO3 | G [auth A] | NITRATE ION N O3 NHNBFGGVMKEFGY-UHFFFAOYSA-N |  | ||
| EOH Query on EOH | O [auth A], P [auth A], R [auth A] | ETHANOL C2 H6 O LFQSCWFLJHTTHZ-UHFFFAOYSA-N |  | ||
| K Query on K | H [auth A] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| CL Query on CL | D [auth A], E [auth A], F [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MOH Query on MOH | N [auth A], Q [auth A], S [auth A] | METHANOL C H4 O OKKJLVBELUTLKV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 129.714 | α = 90 |
| b = 129.714 | β = 90 |
| c = 129.714 | γ = 90 |
| Software Name | Purpose |
|---|---|
| DENZO | data reduction |
| SCALEPACK | data scaling |
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| ProDC | data collection |
| BEAST | phasing |