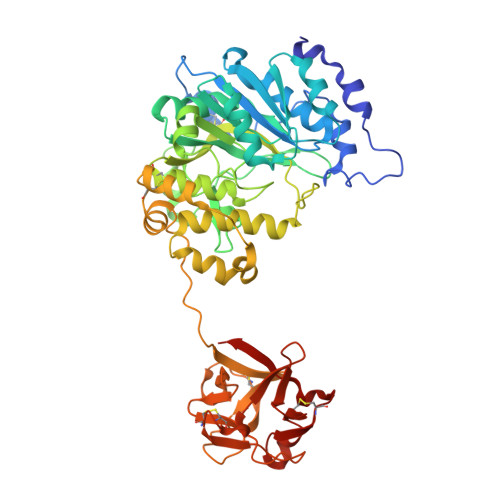
Dynamic Association between the Catalytic and Lectin Domains of Human UDP-GalNAc:Polypeptide {alpha}-N-Acetylgalactosaminyltransferase-2
Fritz, T.A., Raman, J., Tabak, L.A.(2006) J Biological Chem 281: 8613-8619
- PubMed: 16434399
- DOI: https://doi.org/10.1074/jbc.M513590200
- Primary Citation of Related Structures:
2FFU, 2FFV - PubMed Abstract:
The family of UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferases (ppGalNAcTs) is unique among glycosyltransferases, containing both catalytic and lectin domains that we have previously shown to be closely associated. Here we describe the x-ray crystal structures of human ppGalNAcT-2 (hT2) bound to the product UDP at 2.75 A resolution and to UDP and an acceptor peptide substrate EA2 (PTTDSTTPAPTTK) at 1.64 A resolution. The conformations of both UDP and residues Arg362-Ser372 vary greatly between the two structures. In the hT2-UDP-EA2 complex, residues Arg362-Ser373 comprise a loop that forms a lid over UDP, sealing it in the active site, whereas in the hT2-UDP complex this loop is folded back, exposing UDP to bulk solvent. EA2 binds in a shallow groove with threonine 7 positioned consistent with in vitro data showing it to be the preferred site of glycosylation. The relative orientations of the hT2 catalytic and lectin domains differ dramatically from that of murine ppGalNAcT-1 and also vary considerably between the two hT2 complexes. Indeed, in the hT2-UDP-EA2 complex essentially no contact is made between the catalytic and lectin domains except for the peptide bridge between them. Thus, the hT2 structures reveal an unexpected flexibility between the catalytic and lectin domains and suggest a new mechanism used by hT2 to capture glycosylated substrates. Kinetic analysis of hT2 lacking the lectin domain confirmed the importance of this domain in acting on glycopeptide but not peptide substrates. The structure of the hT2-UDP-EA2 complex also resolves long standing questions regarding ppGalNAcT acceptor substrate specificity.
- Section on Biological Chemistry, NIDDK, National Institutes of Health, Bethesda, Maryland 20892, USA.
Organizational Affiliation: