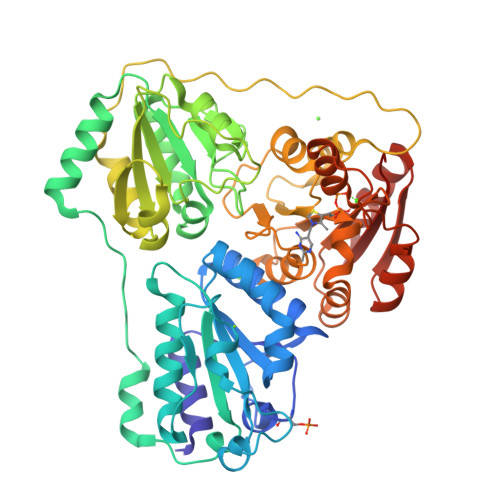
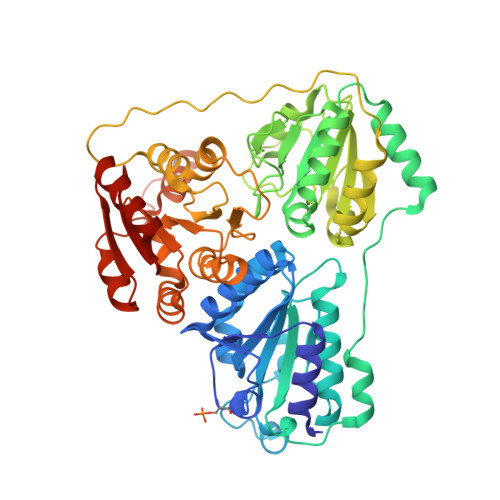
Mechanism-based inactivation of benzoylformate decarboxylase, a thiamin diphosphate-dependent enzyme
Bera, A.K., Polovnikova, L.S., Roestamadji, J., Widlanski, T.S., Kenyon, G.L., McLeish, M.J., Hasson, M.S.(2007) J Am Chem Soc 129: 4120-4121
- PubMed: 17367138
- DOI: https://doi.org/10.1021/ja068636z
- Primary Citation of Related Structures:
2FWN
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907, USA.