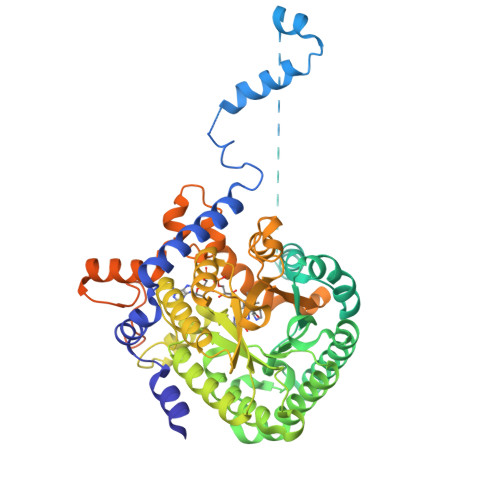
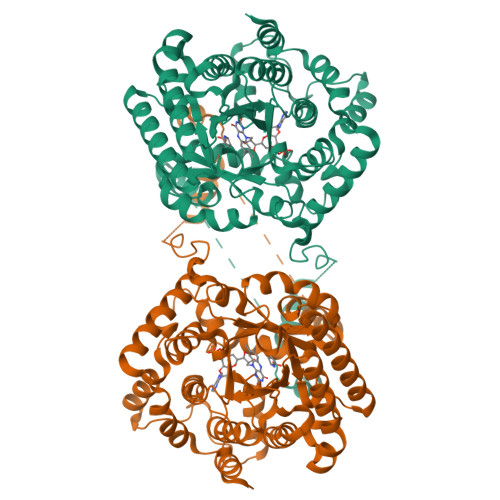
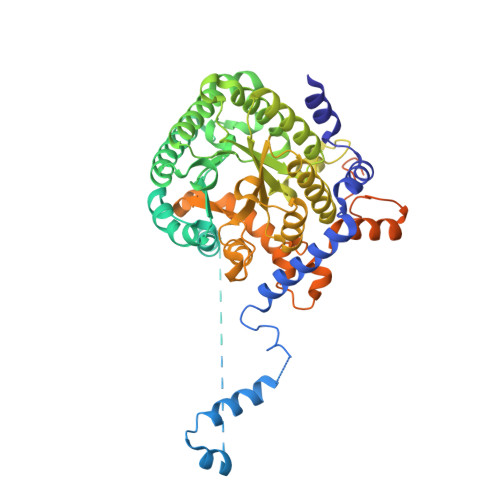
Hydrogen Bonding Interactions of the 2-OH Ribityl Group and the N(5) Position of the FAD Cofactor Regulate PutA-membrane Associations in Escherichia coli
Zhang, W., Zhang, M., Zhu, W., Wanduragula, S., Rewinkel, D., Tanner, J.J., Becker, D.F.To be published.