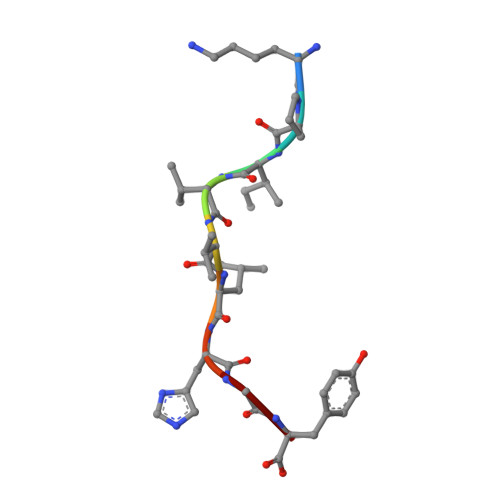
Alloreactivity between disparate cognate and allogeneic pMHC-I complexes is the result of highly focused, peptide-dependent structural mimicry
Archbold, J.K., Macdonald, W.A., Miles, J.J., Brennan, R.M., Kjer-Nielsen, L., McCluskey, J., Burrows, S.R., Rossjohn, J.(2006) J Biological Chem 281: 34324-34332
- PubMed: 16963442
- DOI: https://doi.org/10.1074/jbc.M606755200
- Primary Citation of Related Structures:
2H6P - PubMed Abstract:
Our understanding of the molecular mechanisms of T cell alloreactivity remains limited by the lack of systems for which both the T cell receptor allo- and cognate ligand are known. Here we provide evidence that a single alloreactive T cell receptor interacts with analogous structural regions of its cognate ligand, HLA-B*0801(FLRGRAYGL), as its allogeneic ligand, HLA-B*3501(KPIVVLHGY). The crystal structures of the binary peptide-major histocompatibility complexes show marked differences in the conformation of the heavy chains as well as the bound peptides. Nevertheless, both epitopes possess a prominent solvent-exposed aromatic residue at position 7 flanked by a small glycine at position 8 of the peptide determinant. Moreover, regions of close structural homology between the heavy chains of HLA B8 and HLA B35 coincided with regions that have previously been implicated in "hot spots" of T cell receptor recognition. The avidity of this human T cell receptor was also comparable for the allo- and cognate ligand, consistent with the modes of T cell receptor binding being broadly similar for these complexes. Collectively, it appears that highly focused structural mimicry against a diverse structural background provides a basis for the observed alloreactivity in this system. This cross-reactivity underpins the T cell degeneracy inherent in the limited mature T cell repertoire that must respond to a vast diversity of microbial antigens.
- Protein Crystallography Unit, Department of Biochemistry and Molecular Biology, School of Biomedical Sciences, Monash University, Clayton, Victoria 3800, Australia.
Organizational Affiliation: