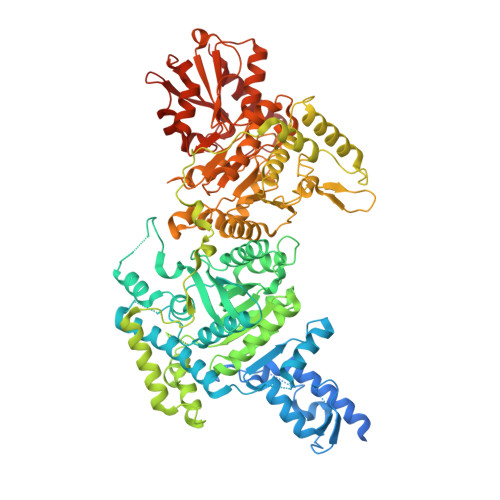
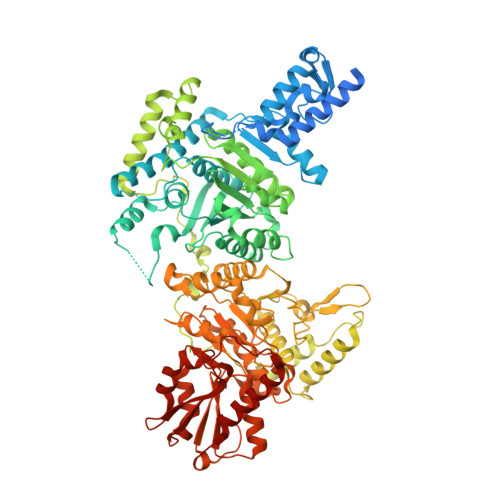
Crystal Structure of the E1 Component of the Escherichia Coli 2-Oxoglutarate Dehydrogenase Multienzyme Complex.
Frank, R.A.W., Price, A.J., Northrop, F.D., Perham, R.N., Luisi, B.F.(2007) J Mol Biology 368: 639
- PubMed: 17367808
- DOI: https://doi.org/10.1016/j.jmb.2007.01.080
- Primary Citation of Related Structures:
2JGD - PubMed Abstract:
The thiamine-dependent E1o component (EC 1.2.4.2) of the 2-oxoglutarate dehydrogenase complex catalyses a rate-limiting step of the tricarboxylic acid cycle (TCA) of aerobically respiring organisms. We describe the crystal structure of Escherichia coli E1o in its apo and holo forms at 2.6 A and 3.5 A resolution, respectively. The structures reveal the characteristic fold that binds thiamine diphosphate and resemble closely the alpha(2)beta(2) hetero-tetrameric E1 components of other 2-oxo acid dehydrogenase complexes, except that in E1o, the alpha and beta subunits are fused as a single polypeptide. The extended segment that links the alpha-like and beta-like domains forms a pocket occupied by AMP, which is recognised specifically. Also distinctive to E1o are N-terminal extensions to the core fold, and which may mediate interactions with other components of the 2-oxoglutarate dehydrogenase multienzyme complex. The active site pocket contains a group of three histidine residues and one serine that appear to confer substrate specificity and the capacity to accommodate the TCA metabolite oxaloacetate. Oxaloacetate inhibits E1o activity at physiological concentrations, and we suggest that the inhibition may allow coordinated activity within the TCA cycle. We discuss the implications for metabolic control in facultative anaerobes, and for energy homeostasis of the mammalian brain.
- Department of Biochemistry, University of Cambridge, 80 Tennis Court Road, Cambridge CB2 1GA, UK.
Organizational Affiliation: