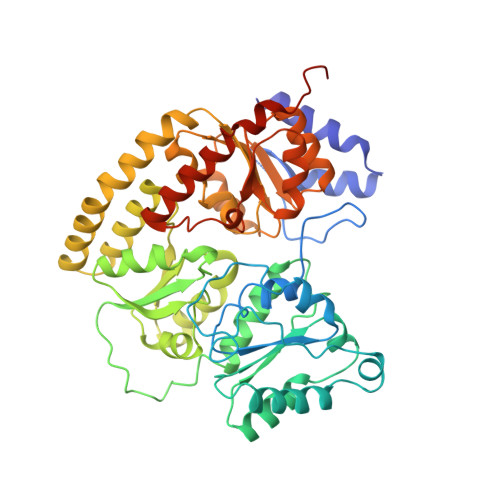
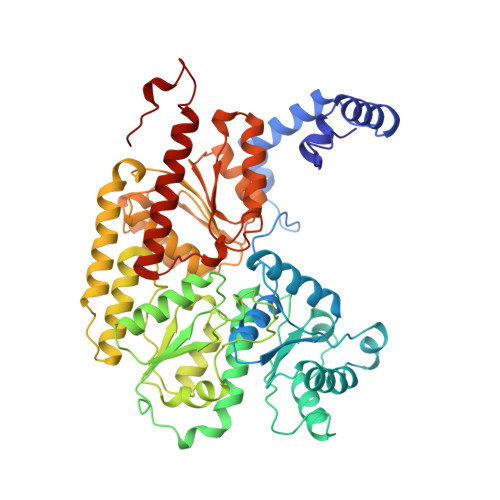
Redox-dependent structural changes in the nitrogenase P-cluster.
Peters, J.W., Stowell, M.H., Soltis, S.M., Finnegan, M.G., Johnson, M.K., Rees, D.C.(1997) Biochemistry 36: 1181-1187
- PubMed: 9063865
- DOI: https://doi.org/10.1021/bi9626665
- Primary Citation of Related Structures:
2MIN, 3MIN - PubMed Abstract:
The structure of the nitrogenase MoFe-protein from Azotobacter vinelandii has been refined to 2.0 A resolution in two oxidation states. EPR studies on the crystals indicate that the structures correspond to the spectroscopically assigned oxidized (P(OX)/M(OX)) and the native or dithionite-reduced (P(N)/M(N)) forms of the enzyme. Both MoFe-protein structures are essentially identical, with the exception of the P-cluster. The MoFe-protein P-cluster in each state is found to contain eight Fe and seven S atoms. Interconversion between the two redox states involves movement of two Fe atoms and an exchange of protein coordination for ligands supplied by a central S atom. In the oxidized P(OX) state, the cluster is coordinated by the protein through six cysteine ligands, Ser-beta188 O gamma, and the backbone amide of Cys-alpha88. In the native P(N) state, Ser-beta188 O gamma and the amide N of Cys-alpha88 no longer coordinate the cluster due to movement of their coordinated Fe atoms toward the central sulfur. Consequently, this central sulfur adopts a distorted octahedral environment with six surrounding Fe atoms. A previously described model of the P-cluster containing 8Fe-8S likely reflects the inappropriate modeling of a single structure to a mixture of these two P-cluster redox states. These observed redox-mediated structural changes of the P-cluster suggest a role for this cluster in coupling electron transfer and proton transfer in nitrogenase.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, Pasadena 91125, USA.
Organizational Affiliation: