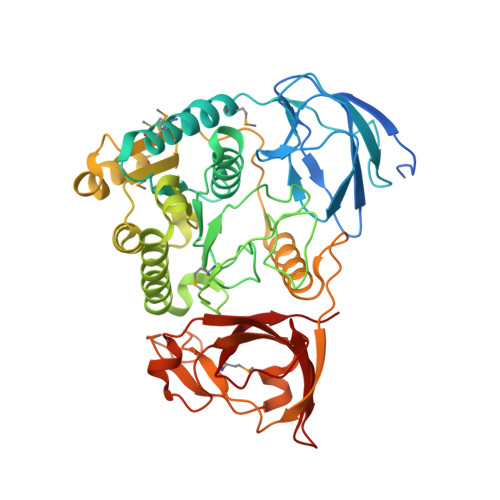
Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor
Noble, C.G., Beuth, B., Taylor, I.A.(2007) Nucleic Acids Res 35: 87-99
- PubMed: 17151076
- DOI: https://doi.org/10.1093/nar/gkl1010
- Primary Citation of Related Structures:
2NPI - PubMed Abstract:
Pcf11 and Clp1 are subunits of cleavage factor IA (CFIA), an essential polyadenylation factor in Saccahromyces cerevisiae. We have determined the structure of a ternary complex of Clp1 together with ATP and the Clp1-binding region of Pcf11. Clp1 contains three domains, a small N-terminal beta sandwich domain, a C-terminal domain containing a novel alpha/beta-fold and a central domain that binds ATP. The arrangement of the nucleotide binding site is similar to that observed in SIMIBI-class ATPase subunits found in other multisubunit macromolecular complexes. However, despite this similarity, nucleotide hydrolysis does not occur. The Pcf11 binding site is also located in the central domain where three highly conserved residues in Pcf11 mediate many of the protein-protein interactions. We propose that this conserved Clp1-Pcf11 interaction is responsible for maintaining a tight coupling between the Clp1 nucleotide binding subunit and the other components of the polyadenylation machinery. Moreover, we suggest that this complex represents a stabilized ATP bound form of Clp1 that requires the participation of other non-CFIA processing factors in order to initiate timely ATP hydrolysis during 3' end processing.
- Division of Molecular Structure, National Institute for Medical Research, The Ridgeway, Mill Hill, London NW7 1AA, UK.
Organizational Affiliation: