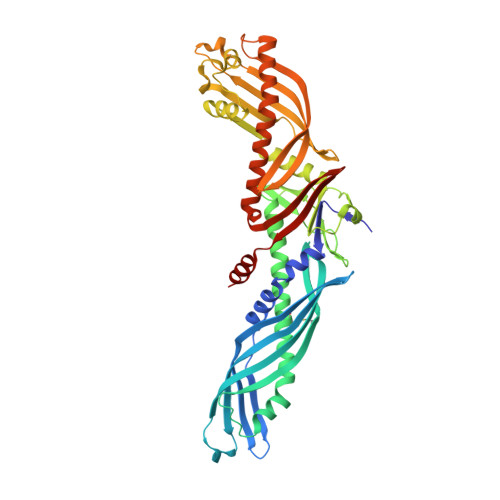
Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Qiu, X., Mistry, A., Ammirati, M.J., Chrunyk, B.A., Clark, R.W., Cong, Y., Culp, J.S., Danley, D.E., Freeman, T.B., Geoghegan, K.F., Griffor, M.C., Hawrylik, S.J., Hayward, C.M., Hensley, P., Hoth, L.R., Karam, G.A., Lira, M.E., Lloyd, D.B., McGrath, K.M., Stutzman-Engwall, K.J., Subashi, A.K., Subashi, T.A., Thompson, J.F., Wang, I.K., Zhao, H., Seddon, A.P.(2007) Nat Struct Mol Biol 14: 106-113
- PubMed: 17237796
- DOI: https://doi.org/10.1038/nsmb1197
- Primary Citation of Related Structures:
2OBD - PubMed Abstract:
Cholesteryl ester transfer protein (CETP) shuttles various lipids between lipoproteins, resulting in the net transfer of cholesteryl esters from atheroprotective, high-density lipoproteins (HDL) to atherogenic, lower-density species. Inhibition of CETP raises HDL cholesterol and may potentially be used to treat cardiovascular disease. Here we describe the structure of CETP at 2.2-A resolution, revealing a 60-A-long tunnel filled with two hydrophobic cholesteryl esters and plugged by an amphiphilic phosphatidylcholine at each end. The two tunnel openings are large enough to allow lipid access, which is aided by a flexible helix and possibly also by a mobile flap. The curvature of the concave surface of CETP matches the radius of curvature of HDL particles, and potential conformational changes may occur to accommodate larger lipoprotein particles. Point mutations blocking the middle of the tunnel abolish lipid-transfer activities, suggesting that neutral lipids pass through this continuous tunnel.
- Pfizer Global Research and Development, Eastern Point Road, Groton, Connecticut 06430, USA. xiayang.qiu@pfizer.com
Organizational Affiliation: