c-Src Binds to the Cancer Drug Imatinib with an Inactive Abl/c-Kit Conformation and a Distributed Thermodynamic Penalty.
Seeliger, M.A., Nagar, B., Frank, F., Cao, X., Henderson, M.N., Kuriyan, J.(2007) Structure 15: 299-311
- PubMed: 17355866
- DOI: https://doi.org/10.1016/j.str.2007.01.015
- Primary Citation of Related Structures:
2OIQ - PubMed Abstract:
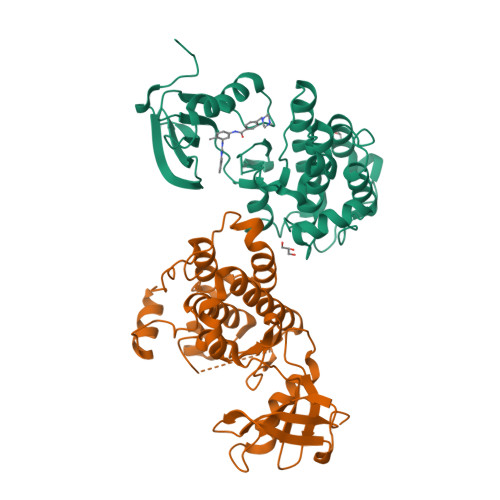
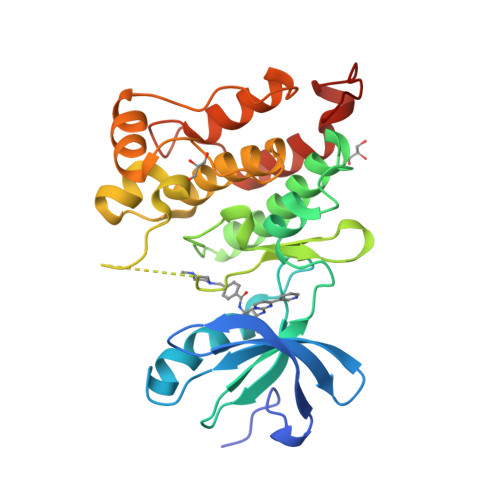
The cancer drug imatinib inhibits the tyrosine kinases c-Abl, c-Kit, and the PDGF receptor. Imatinib is less effective against c-Src, which is difficult to understand because residues interacting with imatinib in crystal structures of Abl and c-Kit are conserved in c-Src. The crystal structure of the c-Src kinase domain in complex with imatinib closely resembles that of Abl*imatinib and c-Kit*imatinib, and differs significantly from the inactive "Src/CDK" conformation of the Src family kinases. Attempts to increase the affinity of c-Src for imatinib by swapping residues with the corresponding residues in Abl have not been successful, suggesting that the thermodynamic penalty for adoption of the imatinib-binding conformation by c-Src is distributed over a broad region of the structure. Two mutations that are expected to destabilize the inactive Src/CDK conformation increase drug sensitivity 15-fold, suggesting that the free-energy balance between different inactive states is a key to imatinib binding.
Organizational Affiliation:
Howard Hughes Medical Institute, University of California, Berkeley, Berkeley, CA 94720, USA.