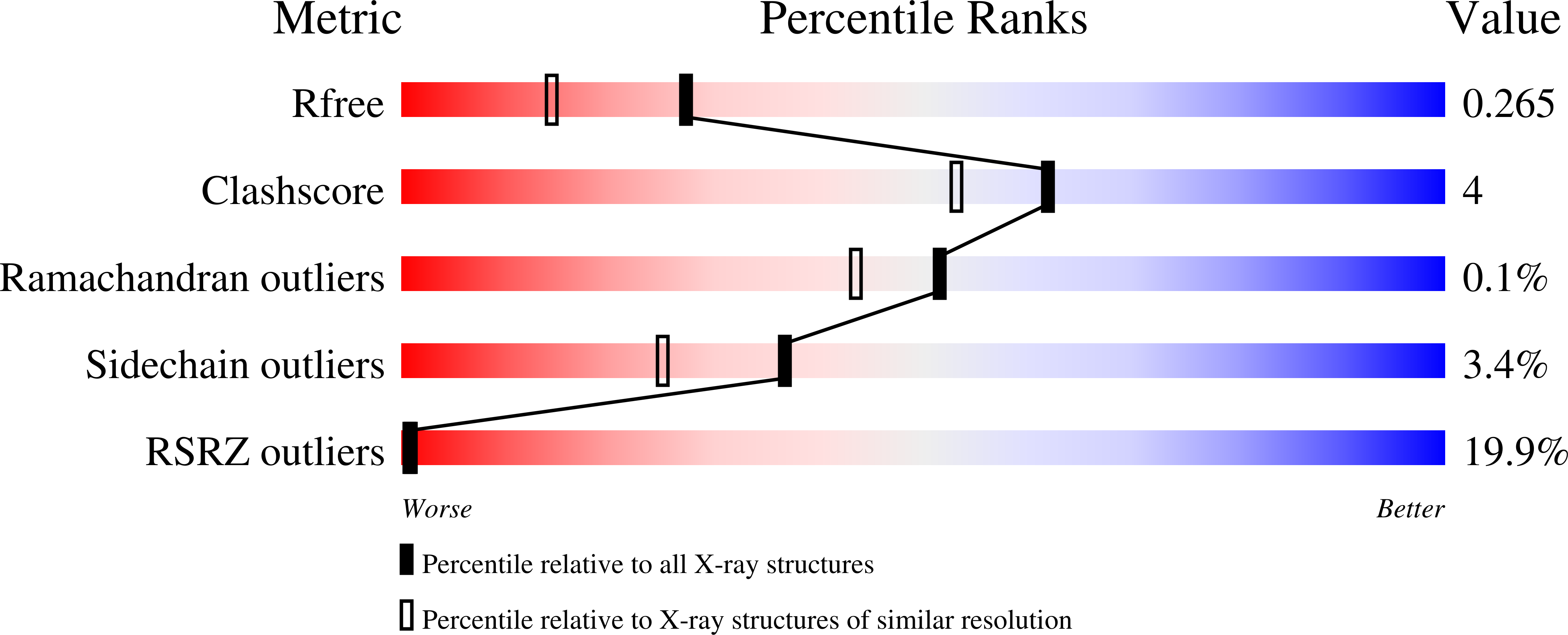
The Crucial Step in Ether Phospholipid Biosynthesis: Structural Basis of a Noncanonical Reaction Associated with a Peroxisomal Disorder.
Razeto, A., Mattiroli, F., Carpanelli, E., Aliverti, A., Pandini, V., Coda, A., Mattevi, A.(2007) Structure 15: 683
- PubMed: 17562315
- DOI: https://doi.org/10.1016/j.str.2007.04.009
- Primary Citation of Related Structures:
2UUU, 2UUV - PubMed Abstract:
Ether phospholipids are essential constituents of eukaryotic cell membranes. Rhizomelic chondrodysplasia punctata type 3 is a severe peroxisomal disorder caused by inborn deficiency of alkyldihydroxyacetonephosphate synthase (ADPS). The enzyme carries out the most characteristic step in ether phospholipid biosynthesis: formation of the ether bond. The crystal structure of ADPS from Dictyostelium discoideum shows a fatty-alcohol molecule bound in a narrow hydrophobic tunnel, specific for aliphatic chains of 16 carbons. Access to the tunnel is controlled by a flexible loop and a gating helix at the protein-membrane interface. Structural and mutagenesis investigations identify a cluster of hydrophilic catalytic residues, including an essential tyrosine, possibly involved in substrate proton abstraction, and the arginine that is mutated in ADPS-deficient patients. We propose that ether bond formation might be orchestrated through a covalent imine intermediate with the flavin, accounting for the noncanonical employment of a flavin cofactor in a nonredox reaction.
Organizational Affiliation:
Dipartimento di Genetica e Microbiologia, Università di Pavia, via Ferrata 1, 27100 Pavia, Italy.