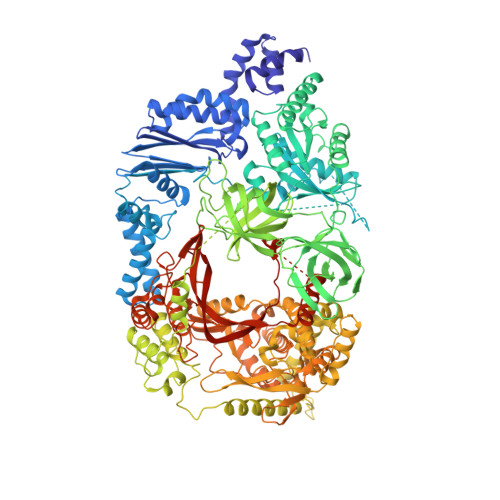
Assembly of Q{beta} viral RNA polymerase with host translational elongation factors EF-Tu and -Ts
Takeshita, D., Tomita, K.(2010) Proc Natl Acad Sci U S A 107: 15733-15738
- PubMed: 20798060
- DOI: https://doi.org/10.1073/pnas.1006559107
- Primary Citation of Related Structures:
3AGP, 3AGQ - PubMed Abstract:
Replication and transcription of viral RNA genomes rely on host-donated proteins. Qbeta virus infects Escherichia coli and replicates and transcribes its own genomic RNA by Qbeta replicase. Qbeta replicase requires the virus-encoded RNA-dependent RNA polymerase (beta-subunit), and the host-donated translational elongation factors EF-Tu and -Ts, as active core subunits for its RNA polymerization activity. Here, we present the crystal structure of the core Qbeta replicase, comprising the beta-subunit, EF-Tu and -Ts. The beta-subunit has a right-handed structure, and the EF-Tu:Ts binary complex maintains the structure of the catalytic core crevasse of the beta-subunit through hydrophobic interactions, between the finger and thumb domains of the beta-subunit and domain-2 of EF-Tu and the coiled-coil motif of EF-Ts, respectively. These hydrophobic interactions are required for the expression and assembly of the Qbeta replicase complex. Thus, EF-Tu and -Ts have chaperone-like functions in the maintenance of the structure of the active Qbeta replicase. Modeling of the template RNA and the growing RNA in the catalytic site of the Qbeta replicase structure also suggests that structural changes of the RNAs and EF-Tu:Ts should accompany processive RNA polymerization and that EF-Tu:Ts in the Qbeta replicase could function to modulate the RNA folding and structure.
- Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology, Higashi, Tsukuba, Ibaraki, Japan.
Organizational Affiliation: